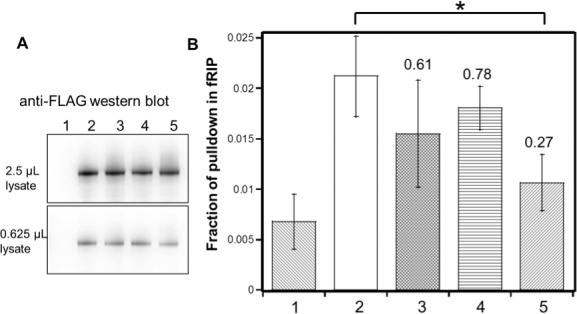
Figure 6. Mutagenesis results support RNA-binding roles of regions Z1, Z3 and Z4 in HDX data, and key residues are identified.
(A) Identification of key residues in Z1. Positions of F32, R34, D36 and K39 in N-terminal helix of EZH2 are indicated in the ac/hsPRC2 crystal structure. K39A point mutant, triple mutant [F32A D36A K39A] and quadruple mutant [F32A R34A D36A K39A] were purified and tested for binding with (GGAA)10 RNA, together with the wild type 3 m hsPRC2. All proteins were used at successive threefold dilutions starting at 2.5 µM concentration. (B) Mutagenesis in Z2, Z3 and Z4 leads to identification of important residues in Z3 and Z4. All proteins were used at successive threefold dilutions starting at 5 µM concentration, and Kdapp values and errors are mean and standard derivation of three binding experiments performed at different days. (C) Histone methyltransferase activity assays for RNA-binding mutants, normalized to that of wild type and error calculated from three independent experiments. Red arrows indicate the mutants that disrupt RNA binding while maintaining normal catalytic activity. (D) Combining identified mutations into a single mutant [F32A D36A K39A and PRKKKR489-494NAAIRS] causes more severe binding defect for (GGAA)10 RNA. Both proteins were used at successive threefold dilutions starting at 5 µM concentration, and Kdapp values and errors are mean and standard derivation of three independent experiments.
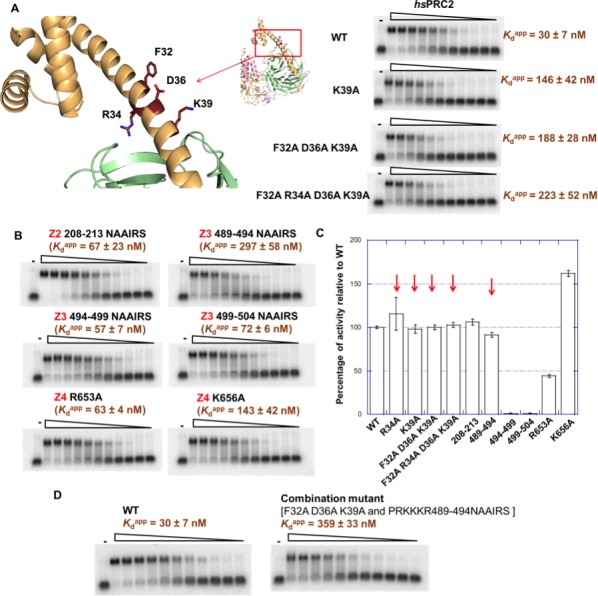
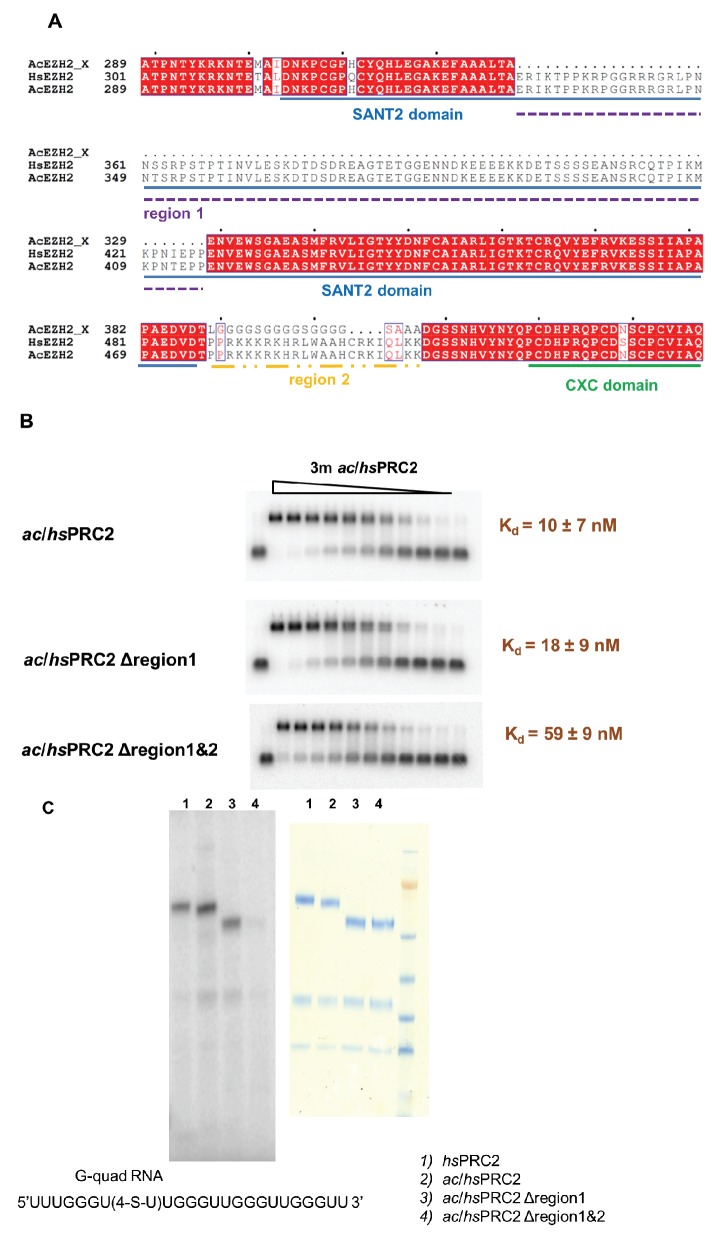
Figure 6—figure supplement 1. A basic region (adjacent to and include Z3 in HDX-MS result) of EZH2 is important for RNA binding.
Figure 6—figure supplement 2. Mutations in the context of the 5 m hsPRC2 (holoenzyme) exhibit binding defect specifically for RNA relative to DNA.
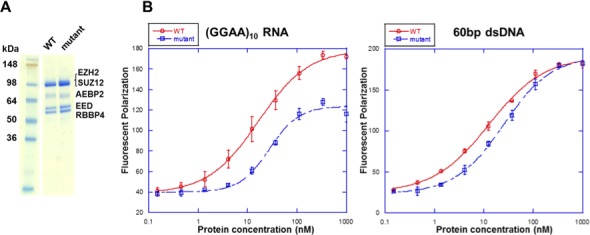
Figure 6—figure supplement 3. Identified residues are important for (GGAA)10 RNA interaction in vivo.