Figure 1. Ppz phosphatase activity is required for proper management of cellular ubiquitin.
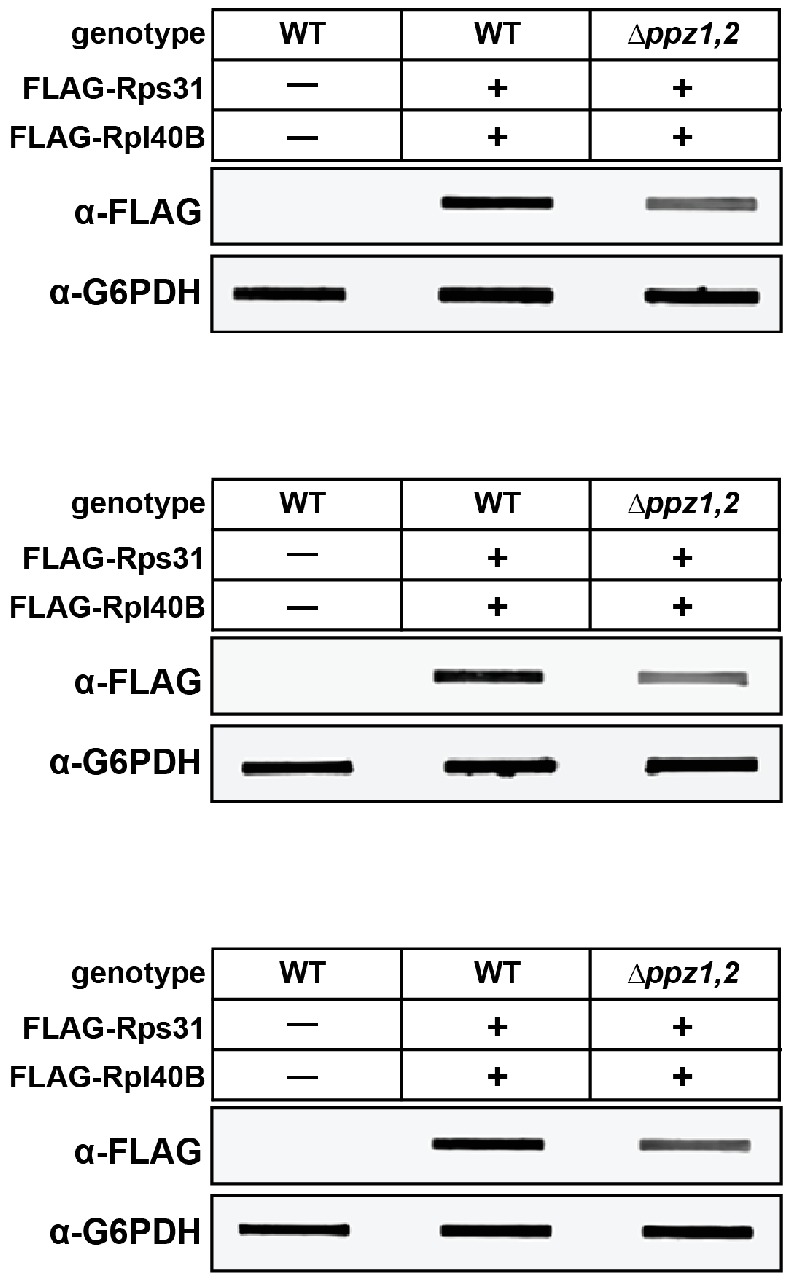
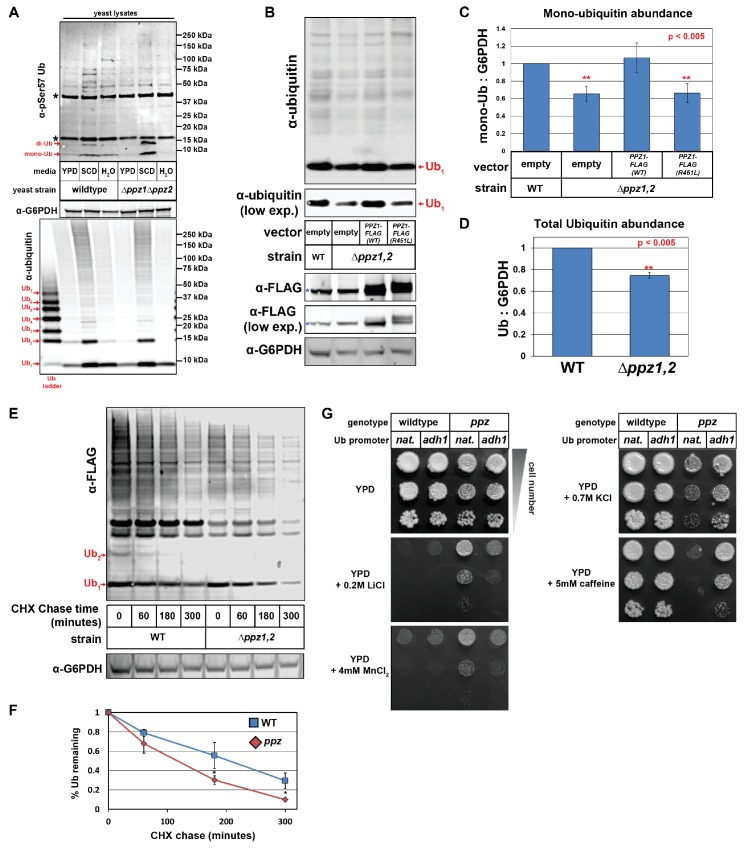
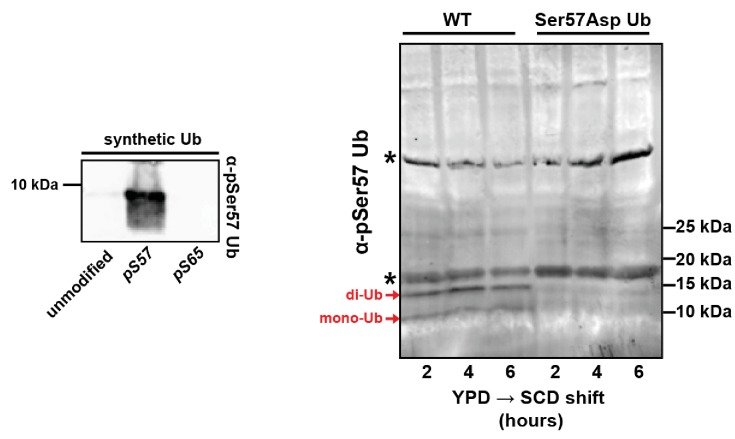
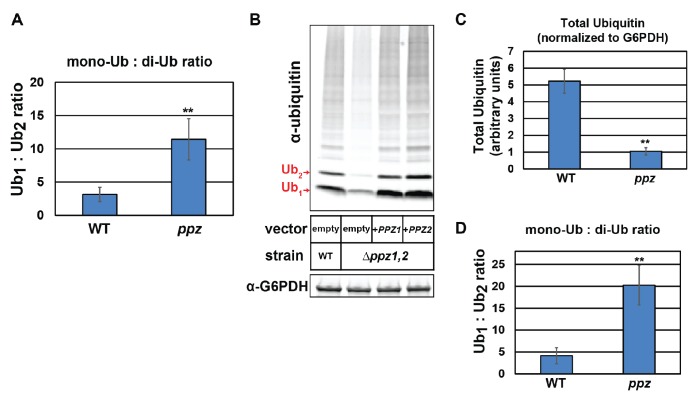
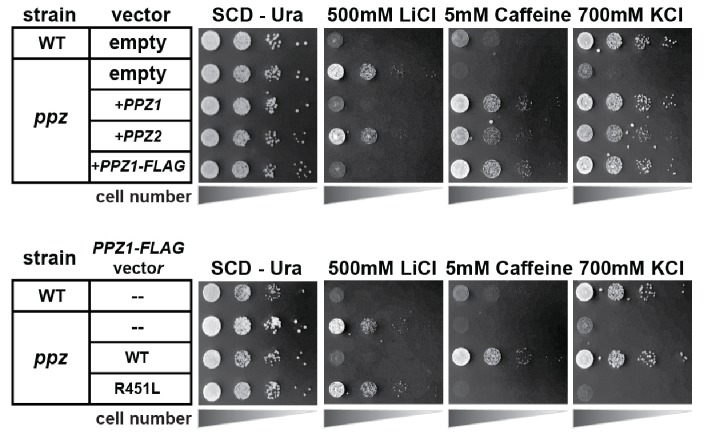
(A) Immunoblot analysis using an α-phospho-Ser57 specific antibody to detect Ser57 phosphorylation of ubiquitin in yeast lysates. Yeast lysates from a SUB280 background, comparing wild-type cells (left three lanes) and ppz mutant cells (right three lanes) grown to mid log phase in YPD (‘YPD’), or shifted from mid log growth in YPD to minimal complete media for 6 hr (‘SCD’) or shifted to growth in water overnight (‘H2O’). Samples were resolved by SDS-PAGE and immunoblotted using antibodies that recognize total ubiquitin (bottom panel), G6PDH (middle panel), or phospho-Ser57 ubiquitin (top panel). For the total ubiquitin blot (bottom panel), a ubiquitin ladder was included as a standard for different unconjugated poly-ubiquitin species (red arrows). For the α–phospho-Ser57 ubiquitin blot (top panel), red arrows indicate species that are specific to Ser57 on ubiquitin and asterisks (*) indicate non-specific bands detected by the antibody, as illustrated in Figure 1—figure supplement 2. (B) The indicated yeast cells (SEY6210 background) containing either empty vector or PPZ1-FLAG vectors were analyzed for total cellular ubiquitin levels by immunoblot analysis. The blue asterisk indicates a background band detectable by FLAG antibody that is a MW similar to Ppz1-FLAG. (C) Quantification of mono-ubiquitin abundance in yeast cell lysates (SEY6210 background) from multiple biological replicates of the immunoblot shown in (B) (n = 5). (D) Total ubiquitin abundance was quantified in wild-type and ppz mutant cells expressing endogenously FLAG-tagged ubiquitin. Total cell lysates were analyzed by quantitative analysis of slot blots shown in Figure 1—figure supplement 3. (E) The indicated yeast cells were grown to mid-log phase and cell lysates were analyzed for total ubiquitin levels at the indicated time points following a cycloheximide (CHX) chase. (F) The results for (F) were quantified over multiple experiments (n = 3). (G) Analysis of yeast growth in the indicated conditions. In this experiment, the yeast ubiquitin gene was expressed from either native (pRPS31) or overexpression (pADH1) promoter was shuffled into the SUB280 strain background. Indicated yeast were plated in 10-fold serial dilutions on indicated plates. In all panels, double asterisk (**) indicates p<0.005 and single asterisk (*) indicates p<0.05.
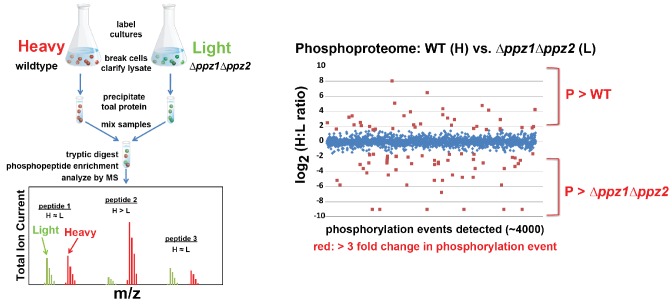
Figure 1—figure supplement 1. A SILAC-based quantitative comparison of the phosphoproteome in wildtype (heavy) and Δppz1Δppz2 (light) cells.
Figure 1—figure supplement 2. Analysis of antibodies recognizing pSer57 ubiquitin.
Figure 1—figure supplement 3. Quantification of cellular ubiquitin levels by analysis of slot blots.