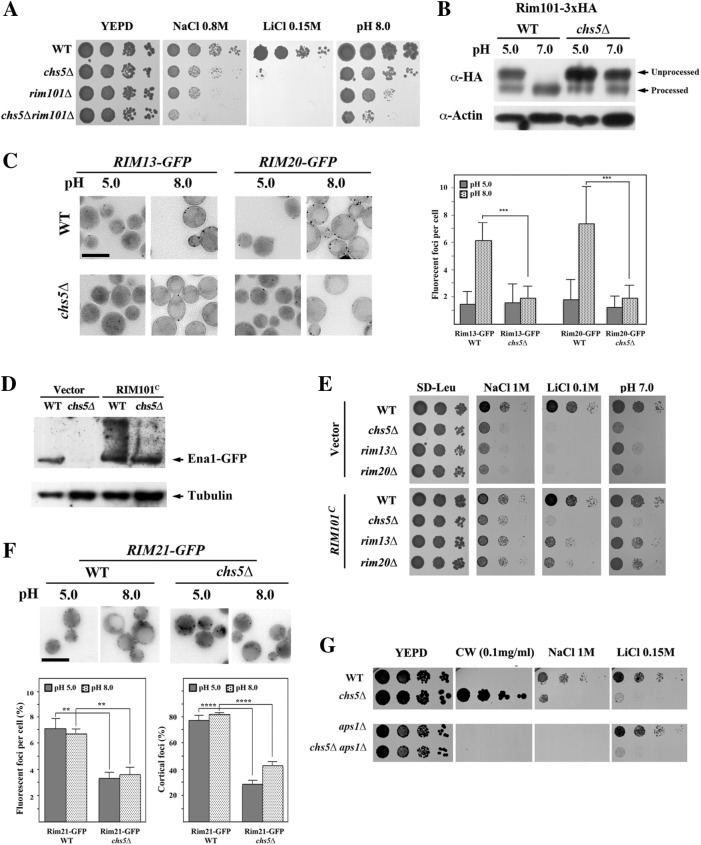
FIGURE 7:
Defective RIM101 signaling in the exomer mutant. (A) Comparative phenotypes of rim101∆ and chs5∆ mutants. Note the similar phenotypes observed and the additive effect of both mutations. (B) Immunoblot of Rim101 proteolytic processing at the indicated pHs. Cells contain a modified version of Rim101 with an internal 3xHA tag. Note the absence of processing in the chs5∆ mutant compared with the control. (C) Visualization of processing spots using yeast cells containing chromosomally tagged versions of Rim13 and Rim20 proteins. Note the increasing numbers of spots for both proteins after alkalinization of the media for 1 h in wild type, which is absent in the chs5∆ mutant. Right panel shows the quantitative results for this experiment, which are the average of three independent experiments. (D) Levels of Ena1-GFP from its endogenous locus after expression of the constitutively processed Rim101C from the pRS315 plasmid. Cells were grown O/N in selective SD media and refreshed in YEPD media for 2 h. (E) Drop assay of strains transformed with the constitutively expressed form of Rim101 (pRS315::RIM101C) on the indicated media. Note the moderate improvement of growth promoted by Rim101C in the chs5∆ mutant under all conditions tested. (F) Rim21-x2GFP localization in the indicated strains/conditions. Note the lower cortical localization of the protein in the chs5∆ mutant independently of the media pH. The quantitative results are the average of at least three independent experiments counting at least 120 cells in each experiment. (G) Phenotypes of the indicated mutants in different media. Note that the absence of Aps1 restores wild-type calcofluor sensitivity of the chs5∆ mutant but not its growth on Na+ or Li+ plates.