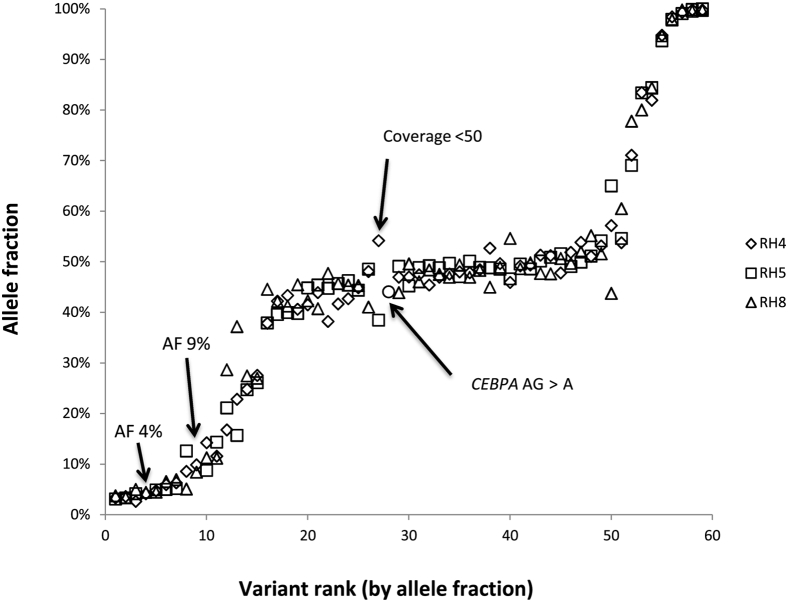
Figure 5.
Inter-run reproducibility. Eight samples comprising 59 single nucleotide variants and insertions/deletions with allele frequencies that ranged from 3% to 100% were tested in three separate runs; among these variants, 55 of 59 (93%) were detected in all three runs. Arrows point to four variants not detected in all three runs. Two variants were in regions with poor coverage in the runs where they were not identified (CEBPA variant was adequately covered in one of three runs and the other had <50× coverage in one run). The two low AF variants (average 4% and 9%) were not identified in a third replicate despite adequate coverage (>200×). Review of the BAM file in Integrated Genome Viewer shows that there were reads of the variant sequences in the third run, but below the pipeline cutoff. AF, allele frequency.