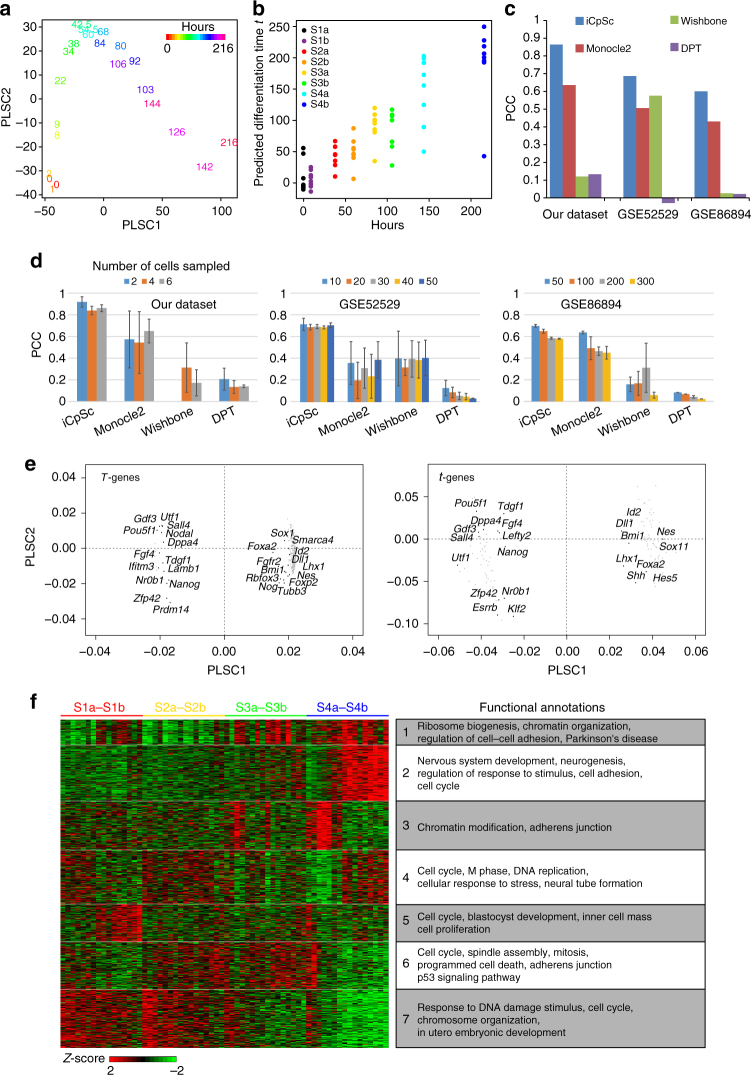
Fig. 2.
Neural differentiation timing in single cells. a Temporal order displayed by cell population samples when projected on the first and second partial least-square (PLS) regression components of cell population RNA-seq profiles. b Single cells ordered by the differentiation time t predicted by the PLSC1 and 2 derived linear model (see Methods). Cells with the same color were obtained at the same experimental time point. c Similarity between real sample collection time and the predicted time and given by four different (pseudo-)time prediction methods on the mESC neural differentiation dataset or two public datasets. d Robustness of four (pseudo-)time prediction methods. Similarity between real sample collection time and the predicted time and given by four different (pseudo-)time prediction methods on the mESC neural differentiation dataset or two public datasets, in randomly selected sample sets with different numbers of cells sampled for each time point. Error bars indicate the standard deviation of five sampling tests. e The projections onto PLSC1 and PLSC2 for selected genes: genes (gray dots) that have the top 10 strongest expression correlations with the fitted differentiation time T in cell population (left) or the predicted differentiation time t in single cells (right), and development-related marker genes that have |PCC| > 0.6 with T or t are shown as black dots with gene names. f The expression pattern of predicted differentiation time-related genes, the “t1–4-genes”, clustered by BIC-Skmeans and their functional annotations