Fig. 3.
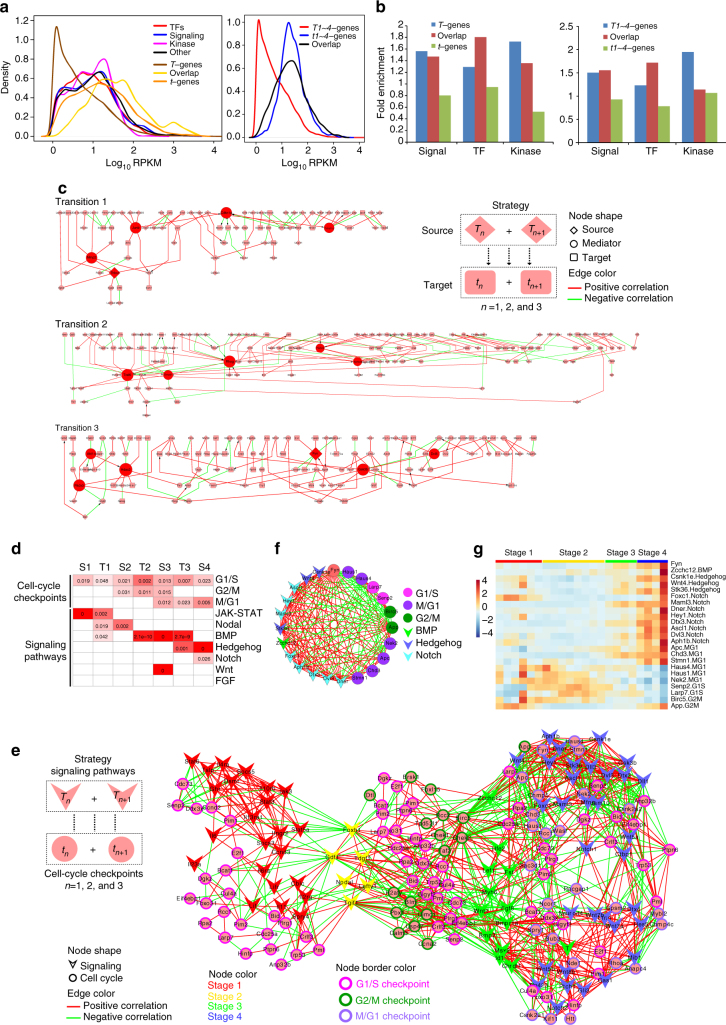
Inferring regulatory events for mESC neural differentiation timing. a Expression level distribution of signaling genes, kinases, and transcription factors, compared with scRNA-seq detectable genes and undetectable cpDEGs (left panel), T-/T1–4-genes, t-/t1–4-genes, or their overlapping genes (right panel) in the cell population RNA-seq data. b Fold enrichment for signaling genes, kinases, and transcription factors in scRNA-seq detectable and undetectable cpDEGs, in T-/T1–4-genes, t-/t1–4-genes, and their overlapping genes. c The largest component of each stage transition eResponseNet. The graphic legend shows the eResponseNet input data. Nodes with degree > 4 (top 5%) are labeled as hubs (large nodes). Red/green edge color represents positive/negative correlation (|PCC| > 0.6) between the two nodes’ expression profiles in the cell population RNA-seq data. See graphic legend for node annotations. d Significance of enrichment for three cell-cycle checkpoints and seven development-related signaling pathways’ targets in four stages and three stage transitions. Significances of enrichment for pathways members are shown in Supplementary Fig. 8c. Significant enrichment is shown by red blocks, with white representing insignificant. Fisher’s exact test was used to test cell-cycle checkpoint enrichment in t n-genes (n = 1, 2, 3, and 4) and in genes of each transition eResponseNet (Bonferroni-corrected p-value < 0.05). GSEA against the rank list sorted by expression levels of all expressed genes, and Fisher’s exact test are used to test signaling pathway enrichment in each stage and in each transition eResponseNet over the whole genome, respectively (GSEA FDR < 0.05, Bonferroni-corrected p-value < 0.05, see Methods section). For column names, “S” stands for “Stage” and “T” for “Transition”. e The CSI network among the T n- or t n-genes belonging to enriched signaling pathways and cell-cycle checkpoints, respectively (n = 1, 2, 3, and 4). Gene expression PCC-derived CSIs are calculated based on cell population RNA-seq expression values. The stage of a gene is defined by the stage where its pathway is activated (see Methods section). f Subnetwork of Fyn from e. Node shapes indicate cell-cycle checkpoints or signaling pathways. Node colors represent different gene categories. g Expression patterns of genes in f network during mESC neural differentiation