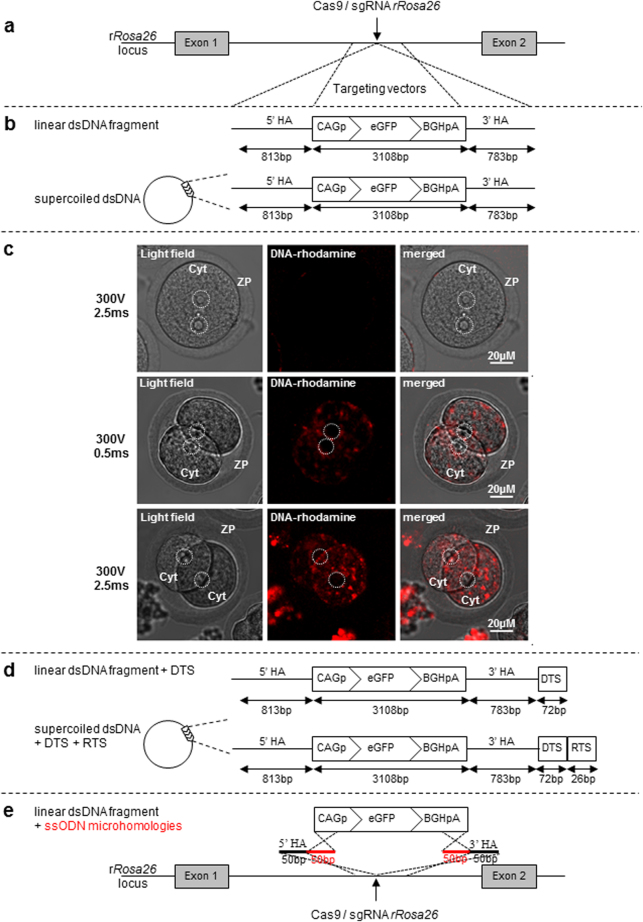
Figure 2.
CRISPR/Cas9 mediated HDR strategies using long dsDNA template. (a) Schematic representation of the rat Rosa26 locus with the site of CRISPRs/Cas9 cleavage (arrow). (b) The targeting vectors containing a CAG-GFP expression cassette (3108 bp) and two homology arms (5′HA 813 bp and 3′ HA 783 bp) contiguous to the cleavage point. These targeting vectors were either in a linear (excised fragment) or supercoiled form. (c) Confocal microscopy images were captured the day after electroporation of rhodamine-labeled linear dsDNA (the same as in b) into intact rat zygotes using a 300 v poring pulse and a pulse width adjusted to 0.5 or 2.5 ms and then cultured overnight. (upper) Embryo electroporated with pulse width of 2.5 ms without incorporated DNA. (middle) Embryo electroporated with pulse width of 0.5 ms. (lower) Embryo electroporated with pulse width of 2.5 ms. ZP, zona pellucida; Cyt, cytoplasm. Pronuclei are indicated by dotted circles. (d) The same targeting vector as in (b) with a DNA targeting sequence (DTS) in a linear form or in a supercoiled form and in this case including Rosa26 targeted sequences (RTS) to be linearized in cellulo by the sgRNA. (e) A linear dsDNA containing the same GFP expression cassette as in (b) but without Rosa26 homology arms. Also depicted, the ssODNs containing 50 bp homologies with Rosa26 sequences at each extremity (in black) and 50 bp homologies with each extremity of the expression cassette (in red).