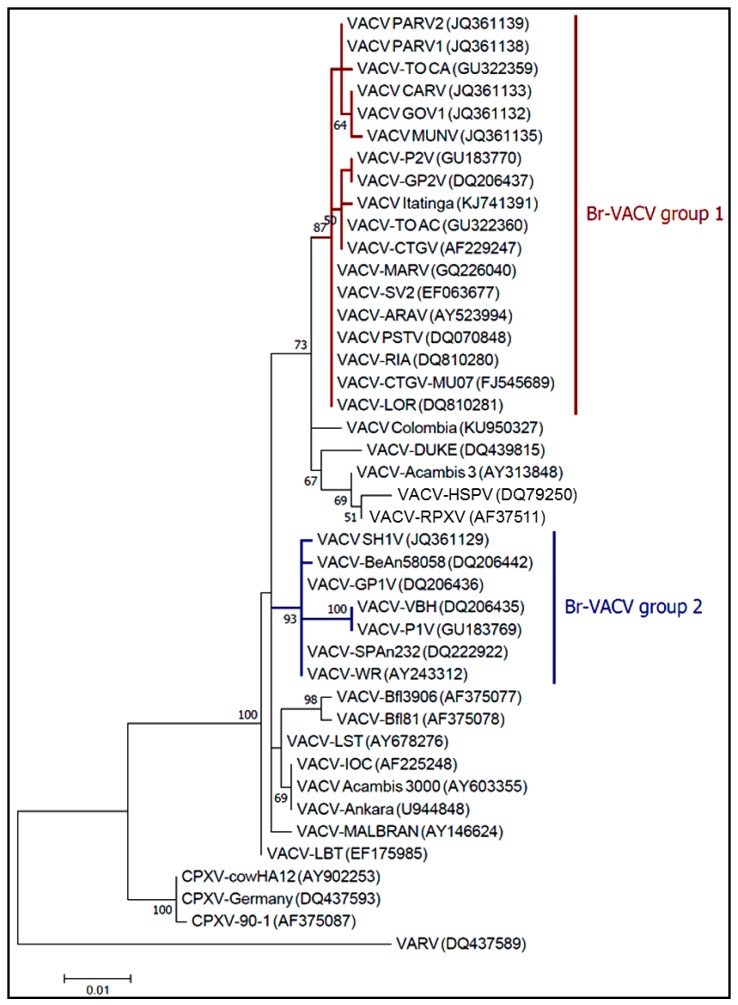
Figure 3.
Phylogenetic analysis based on the A56R gene of VACV vaccine and wild isolates, with cowpox virus (CPXV) sequences included as an outgroup. These sequences are available in the NCBI nucleotide database under the GenBank Accession Numbers provided in brackets on the tree. The sequences were aligned by using ClustalW algorithm and the evolutionary history was inferred by using the Maximum likelihood (ML) method, using Mega 7.0 (GE Healthcare, Buckinghamshire, UK) software and the Jukes-cantor model was selected for ML inference by the program JmodelTest 2.1.6 (Free Software Foundation, Inc., Boston, MA) The evolutionary distances were computed using the Maximum Composite Likelihood method with 1000 Bootstrap replicates. The analysis involved 65 nucleotide sequences with a total of 734 positions in the final dataset. Evolutionary analyses were conducted in Mega 7.0 software.