Abstract
All dsDNA phages encode two proteins involved in host lysis, an endolysin and a holin that target the peptidoglycan and cytoplasmic membrane, respectively. Bacteriophages that infect Gram-negative bacteria encode additional proteins, the spanins, involved in disruption of the outer membrane. Recently, a gene located in the lytic cassette was identified in the genomes of mycobacteriophages, which encodes a protein (LysB) with mycolyl-arabinogalactan esterase activity. Taking in consideration the complex mycobacterial cell envelope that mycobacteriophages encounter during their life cycle, it is valuable to evaluate the role of these proteins in lysis. In the present work, we constructed an Ms6 mutant defective on lysB and showed that Ms6 LysB has an important role in lysis. In the absence of LysB, lysis still occurs but the newly synthesized phage particles are deficiently released to the environment. Using cryo-electron microscopy and tomography to register the changes in the lysis phenotype, we show that at 150 min post-adsorption, mycobacteria cells are incompletely lysed and phage particles are retained inside the cell, while cells infected with Ms6wt are completely lysed. Our results confirm that Ms6 LysB is necessary for an efficient lysis of Mycobacterium smegmatis, acting, similarly to spanins, in the third step of the lysis process.
Keywords: bacteriophage lysis, mycobacteriophage, Ms6, LysB, mycobacteria, spanins, cryo-electron microscopy
1. Introduction
Bacteriophages, the viruses of bacteria, are key elements for biosphere equilibrium, playing a fundamental role in bacterial evolution through constant interactions with their hosts [1,2]. To guarantee their own survival, double-stranded DNA (dsDNA) phages, which represent more than 95% of known bacterial viruses [3], must lyse their hosts. At the end of a lytic cycle, the new phage particles need to be released into the environment, where new host bacteria are potentially available for new infection cycles. The main barrier to phage release is the bacterial cell envelope, and thus, compromising this barrier is the main goal of the lytic process. To accomplish this goal, dsDNA phages synthesize two essential lysis proteins, endolysins and holins. Endolysins are enzymes that disrupt the bacterial cell wall (CW) by cleaving one or more of the five bonds in peptidoglycan (PG). Holins are small proteins that accumulate in the cytoplasmic membrane (CM) and that, at a genetically defined time, form holes in this cell membrane allowing the access of active endolysins to the PG layer or the activation of previously exported endolysins [4,5]. Phages that infect Gram-positive hosts only require the synthesis of these two proteins to compromise the bacterial envelope and consequently for cell burst. However, phages that infect Gram-negative hosts have to face an additional barrier, the outer membrane (OM). It has been shown recently that disruption of this barrier is also required for cell lysis [6]. This is achieved by a third class of lysis proteins named spanins. The best studied spanins are the λ Rz and Rz1 proteins which are an inner membrane and outer membrane protein, respectively. These two proteins form a complex that spans the entire periplasm mediating the fusion of the CM with the OM. This results in the elimination of the last barrier to phage release and consequently, lysis of the host [7,8,9]. Spanin genes, which may encode a sole protein (T1 Gp11) or two subunits like the λ Rz and Rz1 proteins, have been identified in nearly all phages infecting Gram-negative hosts [10,11]. This indicates that, for phages infecting Gram negative hosts, lysis is a three-step event where each component of the cell envelope, i.e., CM, CW and OM is sequentially attacked by holins, endolysins and spanins, respectively [9].
Studies of mycobacteriophage Ms6, a phage that infects Mycobacterium smegmatis, have shown that the lysis cassette composition reflects the complexity of the cell envelope of its host [12]. Although mycobacteria are classified as Gram-positive bacteria, they have a complex cell envelope composed of a CM, similar to other bacterial CMs [13,14], surrounded by a peptidoglycan layer covalently linked to arabinogalactan (AG) which is in turn esterified to a mycolic acid (MA), forming the mycolyl arabinogalactan-peptidoglycan (mAGP) complex [15]. The MAs are long fatty acids that constitute the inner leaflet of a true OM. The outermost leaflet is composed of various glycolipids, including trehalose mono and dimycolate, phospholipids and species-specific lipids [16,17]. Finally, a capsule is composed of proteins, polysaccharides and a few lipids [18,19]. Thus, phages that infect mycobacteria have to overcome this complex envelope for a successful infective cycle. The Ms6 lysis cassette is composed of five genes [20] (Figure 1). In addition to the holin and the endolysin functions, Ms6 encodes a chaperone-like protein (Gp1) that is involved in the delivery of the endolysin to the PG [21,22,23] and an additional lysis protein, Lysin B (LysB), identified as a lipolytic enzyme with the ability to cleave ester bonds of both short and long fatty acids [24]. Experiments with components of the mycobacterial cell envelope showed that Ms6 LysB is a mycolyl-arabinogalactan esterase that cleaves the ester bond between the mycolic acids and the arabinogalactan, and this allows the separation of the OM from the CW [25]. Analogies can be made between Ms6 LysB and the spanins, where Ms6 LysB functions to mediate the final step of host cell lysis.
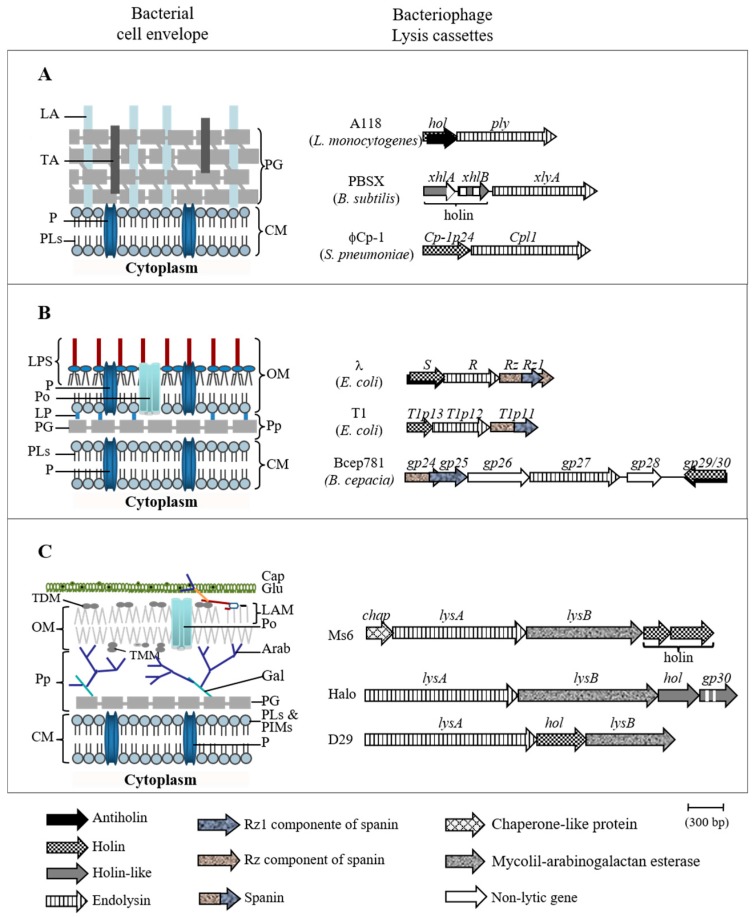
Figure 1.
Cell envelopes of bacteria (left) and representative lysis cassettes of their infecting phages (right). (A) Gram-positive bacteria; (B) Gram-negative bacteria; (C) Mycobacteria. The white segments in holin-like genes indicate the number and position of transmembrane domain coding sequences. Abbreviations: CapGlu, capsular glucan CM, cytoplasmic membrane; LA, lypoteichoic acid; LAM, lipoarabinomannan; LP, lipoprotein; LPS, lipopolysaccharide; P, protein; PG, peptidoglycan; PIMs, phosphatidylinositol mannosides; PLs, phospholipids; PO, porin; Pp, periplasm; TDM, trehalose dimycolate; TMM, trehalose monomycolate. Adapted from reference [5] with permission.
In the present work, we examine the importance of Ms6 LysB in phage lysis and taking advantage of cryo-electron microscopy (cryo-EM) and tomography (cryo-ET), we compare the Ms6 wild-type lysis phenotype with that of a Ms6 mutant lacking the lysB gene. We present evidence that absence of LysB in the Ms6 infection cycle results in incomplete lysis and suggest that the LysB role in lysis parallels that of spanins.
2. Materials and Methods
2.1. Bacterial Strains, Phages, Plasmids and Culture Conditions
Mycobacteria strains, phages, plasmids and oligonucleotides used in this study are listed in Table 1. M. smegmatis strains were propagated in 7H9 medium (BD Biosciences, San Jose, CA, USA) with shaking or Middlebrook 7H10 (BD Biosciences), supplemented with 0.5% glucose, at 37 °C. When appropriate, 1 mM CaCl2 or 15 µg/mL kanamycin was also added to the media. For induced conditions, cells were grown in 7H9 supplemented with 0.2% succinate and induced with 0.2% acetamide.
Table 1.
Bacterial strains, phages, plasmids and oligonucleotides used in this study.
| Name | Description | Source or Reference |
|---|---|---|
| Bacteria | ||
| Mycobacterium smegmatis mc2155 | High-transformation-efficiency mutant of M. smegmatis ATCC 607 | [26] |
| Bacteriophages | ||
| Ms6wt | Temperate bacteriophage from M. smegmatis | [27] |
| Ms6ΔlysB | 996 bp in-frame deletion of the Ms6 lysB gene | This study |
| Plasmids | ||
| pJV53 pAG1 |
Derivative of pLAM12 with Che9c 60 and 61 under control of the acetamidase promoter; Kanr lysB gene cloned into pVVAP |
[28] This study |
| pVVAP | Mycobacteria shuttle vector carrying the acetamidase promoter; Kanr | [29] |
| Oligonucleotides | Sequence 5’-3’ a | |
| PrΔlysB | CTCGGCGGAAAAACCCTCCTCGTGGACGCGGTAGCAGAACTGTTGGGCCACTGATAGGAGGCACCCATGCTGACACGTTCATTCTGGATCGACGCCGCCGAGCG | Ms6ΔlysB |
| PrExtΔlysBFw | CGAGATCCTGCGGCAACTGCGCGGATACAACCTCACTGGCTGGCCGCAGCTCGGCGGAAAAACCCTCGTGGACG | Extend Pr∆lysB |
| PrExtΔlysBRv | CCCCGGCGCCGAGGGTGGCGATCGCGGTTTGGGCGAATGTGCGTATGGCACGCTCGGCGGCGTCGATCCAGAATG | Extend Pr∆lysB |
| PrlysBFw | GCGGATCCATGAGCAGAACTGTTGGGCC | Includes BamHI site to clone in pVVAP |
| PrlysBRv | GGAAGCTTTGTGCGTAGGTAGTCGATG | Includes HindIII site to clone in pVVAP |
| DADA ΔlysB PCRFw | GCGCTAGCAGAACTGTTGGGCCACTGATAG | Ms6ΔlysB |
| DADA Ms6-PCRRv | CGTCTCGTACTGCACGTACCGGTTCTTC | Ms6ΔlysB |
a Restriction sites are underlined.
2.2. Construction of Ms6 Mutant Phage
Construction of Ms6 mutant phage was performed using Bacteriophage Recombineering of Electroporated DNA (BRED) in M. smegmatis as described previously [21,30]. Briefly, for deletion of gene lysB from the Ms6 genome, a 100 bp oligonucleotide (Pr∆lysB), with 50 bp of homology to either flanking region to be deleted was generated. This fragment was extended by PCR to a 200 bp dsDNA substrate using two 75 bp extender primers, PrExt∆lysBFw and PrExt∆lysBRv, sharing 25 bp of homology with either end of the 100-mer. After purification, using MinElute PCR Purification Kit (QIAGEN, Hilden, Germany), the 200 bp substrate was co-electroporated with Ms6wt DNA into electrocompetent recombineering cells of M. smegmatis mc2155:pJV53. Cells were resuspended in 7H9 supplemented with glucose and CaCl2, incubated at 37 °C for 2 h (prior to lysis) and plated on top agar lawns with M. smegmatis mc2155. Individual plaques were recovered and eluted in 100 µL of phage buffer (10 mM Tris-HCl, pH 7.5, 10 mM MgSO4, 68.5 mM NaCl, 1 mM CaCl2), for two hours at room temperature and analyzed by Deletion Amplification Detection Assay (DADA)-PCR [30] with primers DADA ∆lysB-PCRFw/DADA Ms6-PCRRv to detect lysB deletion. Mixed primary plaques containing both wild-type and mutant alleles were eluted as described above, and serial dilutions were plated with M. smegmatis. Individual secondary plaques were screened by DADA-PCR using the same pair of primers.
2.3. Plasmid Construction
To construct plasmid pAG1, a DNA fragment containing the lysB gene was obtained by PCR amplification using Ms6 genomic DNA as template, with primers PrlysBFw/PrlysBRv and Pfu high-fidelity polymerase (Promega®, Madison, WI, USA). Primers were designed in order to have the restriction sites that allow the correct insertion into the shuttle vector pVVAP (V. Visa and M. McNeil; unpublished). All oligonucleotides used were purchased from Thermo Scientific (Waltham, MA, USA) and are listed in Table 1. DNA amplification, plasmid isolation and electrophoresis were carried out using standard techniques [31]. All constructs used in this study were validated and verified by nucleotide sequencing.
2.4. One Step Growth and Single Burst Experiments
One-step growth curves and burst size determination assays [32] were adapted to mycobacteria and carried out in exponential growth phase cell cultures [21]. Briefly, 108 M. smegmatis cells were suspended in 1 mL of a phage suspension at 108 plaque forming units (PFU)/mL. After 50 min of adsorption at 37 °C, nonadsorbed phages were inactivated with 100 µL of 0.4% H2SO4 for 5 min followed by neutralization with 100 µL of 0.4% NaOH. The mixture was diluted 1:100 in 7H9 media and aliquots were taken at intervals of 30 min to quantify the number of phage particles [21]. The obtained results are means of three independent experiments.
A similar procedure was used for burst size determination except that 10 µL of infected cells were diluted in supplemented 7H9 in order to obtain one infected cell/mL. Then, 50 mL of the infected culture was aliquoted into 1 mL volumes and incubated for 3 h at 37 °C. Each sample was plated with 200 µL of M. smegmatis cells and top agar (4 mL) on 7H10 medium and incubated at 37 °C for 24 h. Phage plaques were counted, and the Poisson distribution of (P(n)) was applied to determine the burst size (BS): P(n) = (e−c × cn)/n! (e < 1), where P(n) is the probability of samples having n infected cells, c is the average number of infected cells per tube, and BS (total plaque count in the 50 plates)/(total number of infected cells) [21]. The obtained results are means of three independent experiments.
2.5. Determination of the Number of Phage Particles Released during the Infection Cycle
To determine the number of phage particles released into the supernatant or retained in cells, M. smegmatis was grown up to an OD600 = 0.5, infected with Ms6wt or Ms6ΔlysB at a MOI of 1 and incubated at 37 °C for 3 h. Aliquots were taken at 90 min and 180 min post adsorption and separated by centrifugation into supernatant and pellet fractions. The pellets were suspended in ice-cold phage buffer and sonicated twice for 5 s, with a 30 s interval. Each supernatant and the sonicated pellets were serially diluted using phage buffer and plated on a top agar lawn of M. smegmatis to determine the number of phage particles. Data represent the mean of three independent experiments.
2.6. Cryo-Transmission Electron Microscopy Sample Preparation, Imaging and Image Processing
To observe the lysis phenotype of Ms6wt or Ms6ΔlysB, cells were infected as described above for the one step growth experiment except that the phage input was 100-fold higher. At each time point, 200 µL aliquots were mixed with 10-nm gold nanoparticles (Sigma-Aldrich®, St. Louis, MO, USA). The nanoparticles were later used for image alignment in the 3D tomographic reconstruction process [33,34]. Four µL of the pre-mixed samples were applied to TEM grids that were vitrified by rapid immersion in liquid ethane using a Gatan CryoPlunger3 (Cp3) apparatus (Gatan, Pleasanton, CA, USA). Cryo-grids were transferred to a Gatan 914 high-tilt holder maintained at −178 °C. Cryo-specimens were imaged with JEOL JEM-2200FS 200-kV field emission gun transmission electron microscope (JEOL Ltd., Tokyo, Japan) equipped with an in-column Omega energy filter (slit width 20 eV), a Gatan US4000 4k × 4k CCD camera, and a Direct Electron DE-20 direct detector (Direct Electron, LP, San Diego, CA, USA). Projection images and tilt series were acquired using SerialEM software (http://bio3d.colorado.edu/SerialEM/) [35]. Single-axis tilt series were collected over an angular range of −62° to 62°, with a 2° tilt increment using the DE-20 direct detector. The total electron dose applied to the specimens did not exceed 120 e−/Å2. Tilt series images were acquired at 10,000× nominal magnification (calibrated pixel size of 0.614 nm) with −4 to −8 μm defocus applied. Tomographic reconstructions were generated with IMOD using the r-weighted back-projection algorithm [33,34].
2.7. Nucleotide Sequence Accession Numbers
The phage genome sequences provided in Figure S1 were obtained from GeneBank. The accession numbers are AF022214 for D29, DQ398047 for PBI1; AF319619 for Ms6, DQ398042 for Halo, AY129338 for Omega, GU580941 for ReqiPepy6, GU580940 for ReqiDocB7, KU963246 for SoilAssassin, KX557278 for Ghobes and KR053196 for TPA4.
3. Results
3.1. Ms6 LysB Deletion Decreases Viral Progeny Release
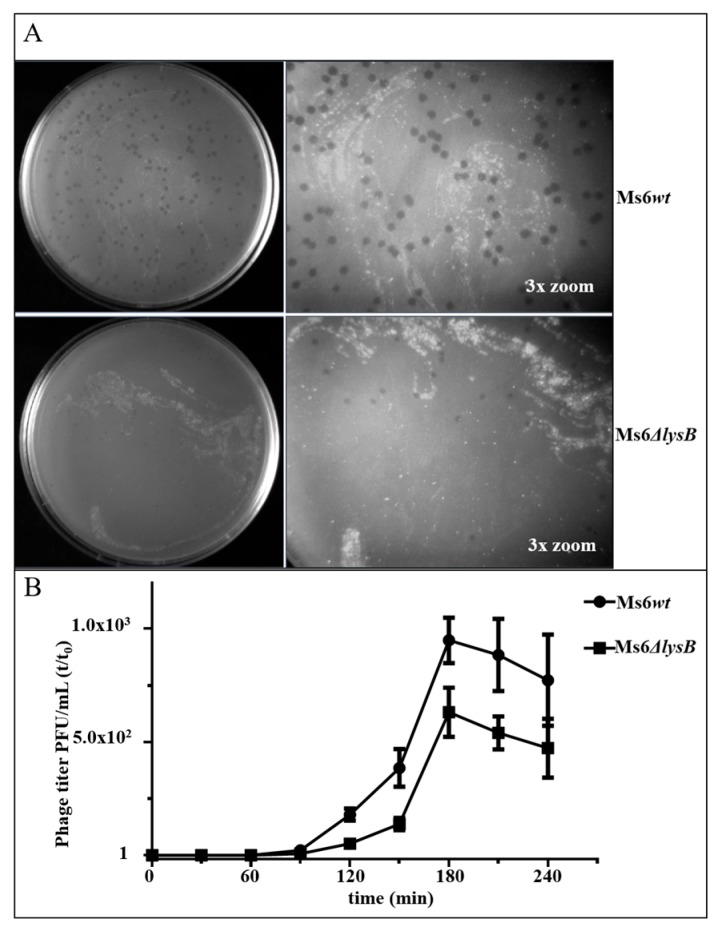
To understand how Ms6 LysB contributes to phage-induced lysis, we took advantage of the Bacteriophage Recombineering of Electroporated DNA (BRED) strategy [30] and constructed an Ms6 derivative mutant lacking gene lysB. The Ms6ΔlysB was able to form plaques on M. smegmatis at equivalent efficiencies to that of the wild-type (wt); however, a reduction in plaque size produced by the mutant was observed (Figure 2A). In a complementation assay, where LysB production was provided from plasmid pAG1, the wild-type phenotype was restored, indicating that plaque size reduction is a consequence of LysB absence.
Figure 2.
(A) Phage plaques formed by Ms6 (top) or Ms6ΔlysB (bottom) on a lawn of M. smegmatis. The plaques formed by Ms6ΔlysB phage are smaller than the ones formed by the wild-type Ms6; (B) one-step growth curves of Ms6wt (circles) or Ms6ΔlysB (squares) on M. smegmatis mc2155 show a lower number of plaque-forming units (PFU) released from Ms6ΔlysB infection. Both curves show similar progression up to 90 min post-adsorption showing no differences in the timing of lysis. T0 marks the end of the adsorption and start of the one-step experiment. The PFU/mL at t = 0 was used to normalize PFU/mL of each time point. For each time point, the mean ± SD of four independent assays is indicated.
To test whether this phenotype results from changes in the phage growth parameters, one-step growth and single-burst experiments were performed. M. smegmatis cells were infected with Ms6wt or Ms6ΔlysB at a multiplicity of infection (MOI) of one. The one-step growth curves (Figure 2B) obtained for Ms6wt and Ms6ΔlysB show that the latent period is similar and that LysB has no effect on the lysis timing; however, the number of infective particles released after Ms6ΔlysB infection was lower than in an Ms6wt infection. Single-burst experiments performed to compare the viable progeny released from single cells show that a Ms6wt infection released an average of 147 ± 27 viable phages per bacterium, while Ms6ΔlysB yielded a reduced burst size of approximately 53 ± 14, where the ± values indicate the mean SD of three independent experiments. Again, when LysB was provided in trans, the wt burst size was restored. These results show that, although Ms6ΔlysB can accomplish lysis of the host cell, the overall process seems to be less efficient.
3.2. Ms6 Is Trapped in Cell Debris in Absence of LysB
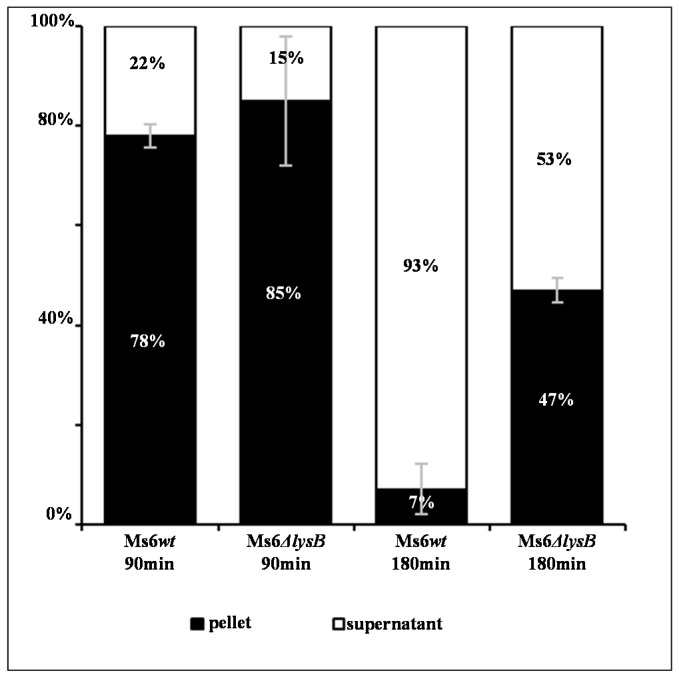
Taking into consideration the observed lysis defect and that: (i) LysB is produced at a late stage of the infection cycle as other lysis proteins, (ii) this protein is a lipolytic enzyme that cleaves the linkage of the mycobacterial OM to the mAGP complex; we hypothesize that the reduced burst size results from a release defect and not from a reduction in the number of synthesized phage particles. To address this question, we performed a time course infection assay with either Ms6wt or Ms6ΔlysB and at each time point the cell pellet was separated from the supernatant and the number of phage particles in each fraction was determined. As observed in Figure 3, at 90 min post-adsorption the majority of the phage particles are not yet released and the number of PFU in the supernatant is similar for both phage infections. However, at 180 min post-adsorption, for the wt phage infection over 90% of phage particles are free in the supernant and only 7% are in the pelleted fraction, while for the Ms6ΔlysB infection, a remarkable 47% of total phage progeny is retained in the pellet. These results confirm that the reduced number of phage particles obtained for the mutant phage, in the single burst experiment, results from a deficient cell lysis, where part of the newly synthesized virions are trapped in incompletely lysed cells.
Figure 3.
Distribution of phage particles in the supernatant and pellet of M. smegmatis infected with Ms6wt or Ms6ΔlysB. Ms6 is trapped in cell debris in absence of LysB. At the indicated time points, the distribution of phage particles in the pellet and in the supernatant was determined as a percentage of the total amount of PFU counted in both fractions. The values indicate the mean ± SD of three independent experiments.
3.3. Cryo-EM Shows Incomplete Cell Lysis in Absence of Ms6 LysB
To prove that the unreleased phage particles remained trapped in incompletely lysed cells, we used cryo-electron microscopy (cryo-EM) and cryo-electron tomography (cryo-ET). This method allows us to visualize the host cell lysis and the viral progeny in their native environment and to examine the lysis behavior of M. smegmatis infected with Ms6wt or Ms6ΔlysB. From each infected M. smegmatis culture, with either Ms6wt or Ms6ΔlysB, aliquots were plunge frozen on copper grids for cryo-EM assessment.
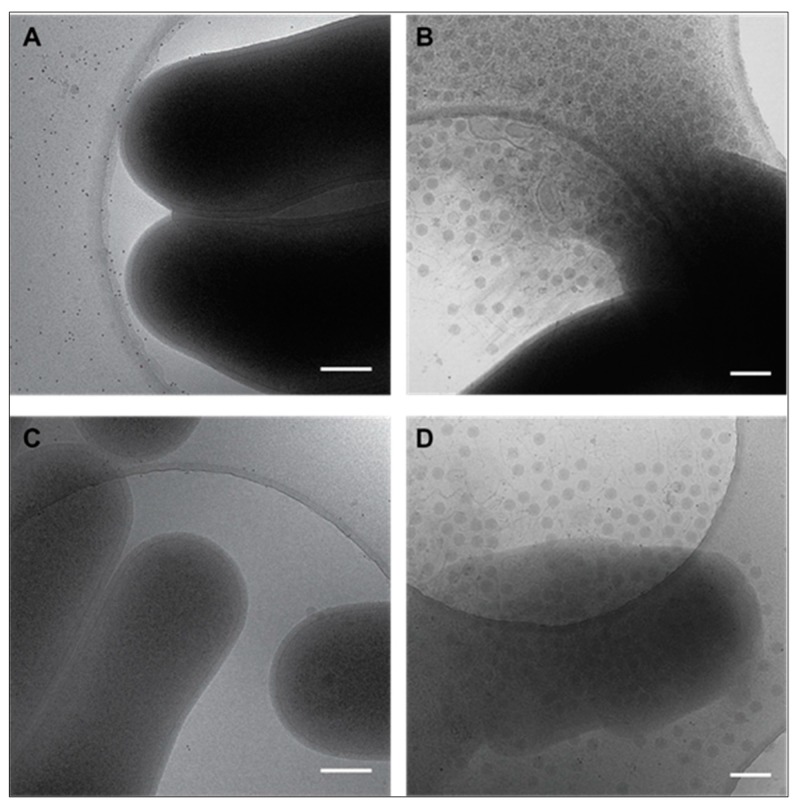
Figure 4 shows collected images of infected cells at 90 and 150 min post-adsorption. At 90 min, no lysis is yet observed (Figure 4A,C). At 150 min post-adsorption, cells infected with Ms6wt burst and release almost all the phages (Figure 4B), while cells infected with Ms6ΔlysB show incomplete lysis and many phages are not released (Figure 4D). Incompletely lysed cells are still captured up to 240 min post-adsorption with the mutant phage, while for the wild-type infection only free phage particles and cell debris are observed.
Figure 4.
Cryo-EM images of M. smegmatis infected with Ms6wt or Ms6ΔlysB. At 90 min post-adsorption, cells infected with Ms6wt (A) or Ms6ΔlysB (C) are still intact and no difference is observed. At 150 min post-adsorption, the abrupt burst of a cell infected with Ms6wt is clear (B) while cells infected with Ms6ΔlysB (D) do not lyse abruptly and deformations in the cell envelope are clearly visible. Scale bar (200 nm).
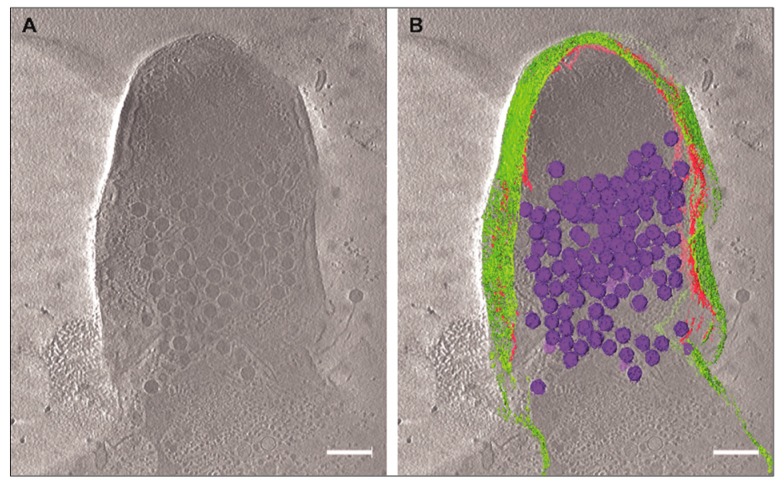
Cryo-ET data collection was performed on Ms6ΔlysB infected cells. In Figure 5A, a central slice through the 3D tomogram shows phages inside the incompletely lysed cell and what appears to be lesions throughout the cell envelope. To facilitate the visualization of the phages and to demonstrate they are inside the cell, segmentations of several 3D tomographic volumes was performed. With this method, it is possible to render the structures or regions of interest in the tomogram (Figure 5B). It is clear that many phages are inside the incompletely lysed cell despite the evident deformation of the cell envelope. It is also clear that most of the CM and PG are absent, while the OM still remains as a veil surrounding and holding some of the cell content.
Figure 5.
Cryo-electron tomography of M. smegmatis infected with Ms6ΔlysB at 150 min post-adsorption. (A) Slice through the tomogram of an infected cell; (B) segmented volume of the phage capsids (purple), cell membrane and PG (magenta) and the outer membrane (green). Scale bar (200 nm).
4. Discussion
It is well known that lysis of the bacterial host is the last event of dsDNA phage lytic cycle, so that the new synthesized phage particles may be released into the environment and infect new available hosts. Compromising the bacterial cell barriers is a sine qua non condition to achieve this final step. Although the role of holins and endolysins has long been well defined, targeting the CM and the CW respectively, the importance of spanins in lysis has only recently been established [9]. The best characterized spanin is that of phage λ, which is composed of two subunits, the Rz and Rz1 proteins, that, once localized to the inner and outer membranes, respectively interact by the C-termini of their periplasmic domains to form a complex that spans the entire periplasm [7,10]. For many years, Rz/Rz1 were considered auxiliary genes, because under laboratory conditions λ lysis could be achieved in the absence of these genes, unless the OM was artificially stabilized by the presence of millimolar concentrations of Ca2+ [36]. Recently, Berry et al. [7,9] have demonstrated that, in nature, in absence of stabilizing cations, these proteins are required for λ lysis, as induction of λ lysogens in the absence of a spanin function results in lysis failure. The infection cycle terminates leading to spherical cells where the CM and PG have been disrupted, but the OM remains intact, indicating that the latter is an important barrier to lysis. In a λ lytic cycle, the Rz-Rz1 complexes accumulate in the envelope during the morphogenesis phase. It was suggested that, following PG disruption by the endolysins, the spanins function by fusing the inner and outer membrane, this results in outer membrane disruption and consequently cell lysis [7,9].
This spherical phenotype in the absence of spanins has also been observed in infections with phages P2 [37] and PRD1 [11]. The presence of Rz/Rz1 equivalents in the lysis cassette, of nearly all bacteriophages that infect Gram-negative hosts, strengthens the idea that to accomplish lysis, in addition to compromising the CM and the CW through the action of holins and endolysins, these phages also need to disrupt the OM [7,10].
In this work, we show that mycobacteriophage Ms6, a phage that infects the mycobacterial species, M. smegmatis, in addition to the holin and endolysin functions also requires an additional lysis protein to overcome the last cell barrier. We provide evidence that Ms6 LysB parallels the function of spanins.
Mycobacteria, which are members of the Corynebacteriales order, are bacteria that, despite being classified as Gram-positive, share a complex cell envelope. In addition to a CM and a CW, they also contain an OM, which is an asymmetrical bilayer where the inner leaflet, composed of long chain mycolic acids, is linked to the CW through an ester bond to AG. This peculiar OM confers to mycobacteria their characteristic impermeability and resistance to therapeutic agents, and as so, is also predicted to be a barrier to mycobacteriophage-induced lysis [13,14,15,16,17]. We have previously shown that Ms6 LysB is a lipolytic enzyme that cuts the linkage between AG and MA on the mycolyl-arabinogalactan-peptidoglycan complex [24,25].
We have observed that, in contrast with Ms6 LysA [23] and in general with phage endolysins, the Ms6 LysB, under our laboratory conditions, is not essential for the phage life cycle, since the Ms6 derivative mutant lacking gene lysB is viable and capable of forming plaques in M. smegmatis. We have observed, however, a reduction in the plaque size. In a one-step assay, we could demonstrate that in absence of LysB there is a defective phage release at the end of an infection cycle. Indeed, in the single burst experiment a reduction of 64% in the number of free phage particles per bacterium was observed. This is in agreement with the reduced plaque size of Ms6ΔlysB, a phenotype that was reverted to the wild-type when LysB was provided in trans. Since Ms6 LysB is produced during late gene expression, from gene lysB, which is part of the lysis cassette, a role in host lysis is obviously expected.
A reduced phage release, together with a reduced plaque size was also reported for the LysB of mycobacteriophage Giles; however, and in contrast to phage Ms6 where absence of LysB did not affect the timing of lysis, the authors observed that lysis induced by GilesΔlysB was delayed in 30 min when compared to the Gileswt [38]. It is not clear so far how the absence of Giles LysB affects the timing of lysis.
The observation that, at the end of a Ms6ΔlysB infection, 47% of the phages are recovered from the cell pellet against only 7% in a Ms6wt infection, indicates that the reduced burst size results from a deficient phage release and not from a reduction in the number of new synthesized phage particles. In the absence of LysB, phage particles are trapped in incompletely lysed cells. Cryo-EM and cryo-ET of infections in the absence of LysB clearly show unreleased phage particles inside cells infected with Ms6ΔlysB, while at the same time point (150 min post-adsorption) in a Ms6wt infection the cell completely bursts. The 3D tomogram (Movie S1) also shows deformations of the cell, indicating that the OM still holds part of the cell content even after disruption of the CM and PG (Figure 5B), following holin and endolysin action.
Our results support the notion that the role of Ms6 LysB in lysis equates to that of spanins, however with different modes of action, since the structure of mycobacteria OM is completely different from that of Gram-negative bacteria. While spanins function either as a complex (λ Rz-Rz1) or as a single protein (T1 Gp11) by fusing the CM and OM [39], Ms6 LysB protein functions as an enzyme that detaches the OM from the CW by cleaving the bond that links these two structures. As a lipolytic enzyme, Ms6 LysB also acts as an esterase on other lipids containing mycolic acids, such as the trehalose dimycolate (TDM) [25], a glycolipid with an important role in M. tuberculosis pathogenesis [40]. However, it is unknown if cleavage of these lipids contributes to lysis.
The fact that the vast majority of mycobacteriophages sequenced so far encode Ms6 LysB homologous proteins suggests that they have an important role in nature. This is also true for other phages that infect members of the mycolata group, a bacterial group that also contain a layer of mycolic acid-containing lipids in their envelope. Examples are the Rhodococcus equi phages ReqiDocB7, ReqiPepy6 and ReqiPoco6, which encode Ms6 LysB homologues [12,41]. A huge number of genome sequences from phages infecting the same bacterial group is available at The Actinobacteriophage Database (http://phagesdb.org/), and here we can also find genes from several phages annotated as coding for Lysin B as exemplified by gp24 or gp41 from phages SoilAssassin and Ghobes, respectively, both infecting Gordonae terrae [42]. In other cases, although no LysB annotation exists, we could identify the GXSXG motif common to lipolytic enzymes in the deduced amino acid sequence of several genes, such as gp54 from the TPA4 phage, a lytic phage that infects Tsukamurella species (Figure S1).
Collectively our results lead to the suggestion that mycobacteriophage-induced lysis is also a three-step process where holins subvert the cytoplasmic membrane followed by endolysins targeting the cell wall and LysB proteins disrupting the last barrier to mycobacteriophage release, the outer membrane.
Our present knowledge of the mechanism of bacteriophage lysis suggests that the complexity of phage lytic cassettes depends on their hosts. Hosts with a simpler envelope, like Gram-positive bacteria, require the phage to possess a simple lytic cassette, with genes encoding proteins targeting the CM and the PG. For bacteria with a more complex envelope that also contain an OM, degradation of the cell wall is necessary but not sufficient for lysis and phages need to produce specific proteins to overcome this barrier. Thus, phages that infect Gram-negative hosts or mycobacteria, in addition to holins and endolysins, synthesize spanins or lipolytic enzymes, respectively (Figure 1).
Acknowledgments
We would like to thank Varalakshmi Vissa and Michael McNeil (Colorado State University, Fort Collins, CO, USA) for providing pVVAP vector and the Robert P. Apkarian Integrated Electron Microscopy Core, Emory University for microscopy services and support. This work was supported in part by Fundação para a Ciência e Tecnologia (FCT-MCES, Portugal) Grant PTDC/IMI-MIC/0694/2012 to MP; Emory University, Children’s Healthcare of Atlanta, and the Georgia Research Alliance to E.R.W.; the Center for AIDS Research at Emory University (P30 AI050409); public health service grant GM104540 to E.R.W. from the NIH/NIGMS, and NSF grant 0923395 to E.R.W. AG (SFRH/BD/87685/2012) is a recipient PhD fellowship from FCT-MCES, Portugal.
Supplementary Material
The following are available online at www.mdpi.com/1999-4915/9/11/343/s1.
Author Contributions
Madalena Pimentel, José Moniz-Pereira and Elizabeth R. Wright conceived and designed the experiments; Adriano M. Gigante, Cheri M. Hampton, Rebecca S. Dillard, Filipa Gil and Maria João Catalão performed the experiments; Madalena Pimentel, Adriano M. Gigante, Elizabeth R. Wright, Cheri M. Hampton and José Moniz-Pereira analyzed the data; Madalena Pimentel wrote and Adriano M. Gigante, Cheri M. Hampton and Elizabeth R. Wright contributed to the writing of the paper.
Conflicts of Interest
The authors declare no conflict of interest. The founding sponsors had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- 1.Rodriguez-Valera F., Martin-Cuadrado A.-B., Rodriguez-Brito B., Pasić L., Thingstad T.F., Rohwer F., Mira A. Explaining microbial population genomics through phage predation. Nat. Rev. Microbiol. 2009;7:828–836. doi: 10.1038/nrmicro2235. [DOI] [PubMed] [Google Scholar]
- 2.Hatfull G.F., Hendrix R.W. Bacteriophages and their genomes. Curr. Opin. Virol. 2011;1:298–303. doi: 10.1016/j.coviro.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ackermann H.W., Prangishvili D. Prokaryote viruses studied by electron microscopy. Arch. Virol. 2012;157:1843–1849. doi: 10.1007/s00705-012-1383-y. [DOI] [PubMed] [Google Scholar]
- 4.Young R., Wang I.N. Phage lysis. In: Calendar R., editor. The Bacteriophages. Oxford University Press; New York, NY, USA: 2006. pp. 104–125. [Google Scholar]
- 5.Catalão M.J., Gil F., Moniz-Pereira J., São-José C., Pimentel M. Diversity in bacterial lysis systems: Bacteriophages show the way. FEMS Microbiol. Rev. 2013;37:554–571. doi: 10.1111/1574-6976.12006. [DOI] [PubMed] [Google Scholar]
- 6.Rajaure M., Berry J., Kongari R., Cahill J., Young R. Membrane fusion during phage lysis. Proc. Natl. Acad. Sci. USA. 2015;112:5497–5502. doi: 10.1073/pnas.1420588112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berry J., Summer E.J., Struck D.K., Young R. The final step in the phage infection cycle: The Rz and Rz1 lysis proteins link the inner and outer membranes. Mol. Microbiol. 2008;70:341–351. doi: 10.1111/j.1365-2958.2008.06408.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berry J., Savva C., Holzenburg A., Young R. The lambda spanin components Rz and Rz1 undergo tertiary and quaternary rearrangements upon complex formation. Protein Sci. 2010;19:1967–1977. doi: 10.1002/pro.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berry J., Rajaure M., Pang T., Young R. The spanin complex is essential for lambda lysis. J. Bacteriol. 2012;194:5667–5674. doi: 10.1128/JB.01245-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Summer E.J., Berry J., Tran T.A.T., Niu L., Struck D.K., Young R. Rz/Rz1 Lysis Gene Equivalents in Phages of Gram-negative Hosts. J. Mol. Biol. 2007;373:1098–1112. doi: 10.1016/j.jmb.2007.08.045. [DOI] [PubMed] [Google Scholar]
- 11.Krupovič M., Cvirkaitė-Krupovič V., Bamford D.H. Identification and functional analysis of the Rz/Rz1-like accessory lysis genes in the membrane-containing bacteriophage PRD1. Mol. Microbiol. 2008;68:492–503. doi: 10.1111/j.1365-2958.2008.06165.x. [DOI] [PubMed] [Google Scholar]
- 12.Pimentel M. Genetics of Phage Lysis. Microbiol. Spectr. 2014;2:1–13. doi: 10.1128/microbiolspec.MGM2-0017-2013. [DOI] [PubMed] [Google Scholar]
- 13.Mamadou D., Marie-Ange D., Nicole G. The cell envelope of Mycobacterium smegmatis: Cytochemistry and architectural implications. FEMS Microbiol. Lett. 1989;52:89–93. doi: 10.1111/j.1574-6968.1989.tb03558.x. [DOI] [PubMed] [Google Scholar]
- 14.Daffé M. The Global Architecture of the Mycobacterial Cell Envelope. In: Daffé M., Reyrat J., Avenir G., editors. The Mycobacterial Cell Envelope. ASM Press; Washington, DC, USA: 2008. pp. 3–11. [Google Scholar]
- 15.Brennan P. Structure, function, and biogenesis of the cell wall of Mycobacterium tuberculosis. Tuberculosis. 2003;83:91–97. doi: 10.1016/S1472-9792(02)00089-6. [DOI] [PubMed] [Google Scholar]
- 16.Hoffmann C., Leis A., Niederweis M., Plitzko J.M., Engelhardt H. Disclosure of the mycobacterial outer membrane: Cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc. Natl. Acad. Sci. USA. 2008;105:3963–3967. doi: 10.1073/pnas.0709530105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zuber B., Chami M., Houssin C., Dubochet J., Griffiths G., Daffe M. Direct Visualization of the Outer Membrane of Native Mycobacteria and Corynebacteria. J. Bacteriol. 2008;190:5672–5680. doi: 10.1128/JB.01919-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lemassu A., Ortalo-Magné A., Bardou F., Silve G., Laneélle M.A., Daffé M. Extracellular and surface-exposed polysaccharides of non-tuberculous mycobacteria. Microbiology. 1996;142:1513–1520. doi: 10.1099/13500872-142-6-1513. [DOI] [PubMed] [Google Scholar]
- 19.Sani M., Houben E.N.G., Geurtsen J., Pierson J., de Punder K., van Zon M., Wever B., Piersma S.R., Jiménez C.R., Daffé M., et al. Direct visualization by Cryo-EM of the mycobacterial capsular layer: A labile structure containing ESX-1-secreted proteins. PLoS Pathog. 2010;6:e1000794. doi: 10.1371/journal.ppat.1000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia M., Pimentel M., Moniz-Pereira J. Expression of mycobacteriophage Ms6 lysis genes is driven by two σ70-like promoters and is dependent on a transcription termination signal present in the leader RNA. J. Bacteriol. 2002;184:3034–3043. doi: 10.1128/JB.184.11.3034-3043.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Catalão M.J., Gil F., Moniz-Pereira J., Pimentel M. The mycobacteriophage Ms6 encodes a chaperone-like protein involved in the endolysin delivery to the peptidoglycan. Mol. Microbiol. 2010;77:672–686. doi: 10.1111/j.1365-2958.2010.07239.x. [DOI] [PubMed] [Google Scholar]
- 22.Catalão M.J., Gil F., Moniz-Pereira J., Pimentel M. The endolysin-binding domain encompasses the N-terminal region of the mycobacteriophage Ms6 Gp1 chaperone. J. Bacteriol. 2011;193:5002–5006. doi: 10.1128/JB.00380-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Catalão M.J., Milho C., Gil F., Moniz-Pereira J., Pimentel M. A second endolysin gene is fully embedded in-frame with the lysA gene of mycobacteriophage Ms6. PLoS ONE. 2011;6:e20515. doi: 10.1371/journal.pone.0020515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gil F., Catalão M.J., Moniz-Pereira J., Leandro P., McNeil M., Pimentel M. The lytic cassette of mycobacteriophage Ms6 encodes an enzyme with lipolytic activity. Microbiology. 2008;154:1364–1371. doi: 10.1099/mic.0.2007/014621-0. [DOI] [PubMed] [Google Scholar]
- 25.Gil F., Grzegorzewicz A.E., Catalão M.J., Vital J., McNeil M.R., Pimentel M. Mycobacteriophage Ms6 LysB specifically targets the outer membrane of Mycobacterium smegmatis. Microbiology. 2010;156:1497–1504. doi: 10.1099/mic.0.032821-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Snapper S.B., Melton R.E., Mustafa S., Kieser T., Jacobs W.R., Jr. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 27.Portugal I., Anes E., Moniz-Pereira J. Temperate mycobacteriophage from M. smegmatis. Acta Leprol. 1989;1:243–244. [PubMed] [Google Scholar]
- 28.Van Kessel J.C., Hatfull G.F. Recombineering in Mycobacterium tuberculosis. Nat. Methods. 2007;4:147–152. doi: 10.1038/nmeth996. [DOI] [PubMed] [Google Scholar]
- 29.Vissa V., McNeil M. (Department of Microbiology, Immunology and Pathology, Colorado State University, Fort Collins; CO 80523, USA). pVVAp is a mycobacteria shuttle vector, containing the pAL5000 replication origin, the kanamycin resistance gene and the acetamidase promoter region. 2008.
- 30.Marinelli L.J., Piuri M., Swigoňová Z., Balachandran A., Oldfield L.M., van Kessel J.C., Hatfull G.F. BRED: A simple and powerful tool for constructing mutant and recombinant bacteriophage genomes. PLoS ONE. 2008;3:e3957. doi: 10.1371/journal.pone.0003957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambrook J., Russell D.W. Molecular cloning: A laboratory manual. 3rd ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY, USA: 2001. [Google Scholar]
- 32.Adams M.H. Methods of study of bacterial viruses. In: Adams M.H., editor. Bacteriophages. Interscience Publishers, Inc.; New York, NY, USA: 1959. pp. 443–522. [Google Scholar]
- 33.Kremer J.R., Mastronarde D.N., McIntosh J.R. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 34.Mastronarde D.N. Dual-axis tomography: An approach with alignment methods that preserve resolution. J. Struct. Biol. 1997;120:343–352. doi: 10.1006/jsbi.1997.3919. [DOI] [PubMed] [Google Scholar]
- 35.Mastronarde D.N. Automated electron microscope tomography using robust prediction of specimen movements. J. Struct. Biol. 2005;152:36–51. doi: 10.1016/j.jsb.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 36.Zhang N., Young R. Complementation and characterization of the nested Rz and Rz1 reading frames in the genome of bacteriophage λ. Mol. Gen. Genet. MGG. 1999;262:659–667. doi: 10.1007/s004380051128. [DOI] [PubMed] [Google Scholar]
- 37.Markov D., Christie G.E., Sauer B., Calendar R., Park T., Young R., Severinov K. P2 growth restriction on an rpoC mutant is suppressed by alleles of the Rz1 homolog lysC. J. Bacteriol. 2004;186:4628–4637. doi: 10.1128/JB.186.14.4628-4637.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Payne K., Sun Q., Sacchettini J., Hatfull G.F. Mycobacteriophage lysin B is a novel mycolylarabinogalactan esterase. Mol. Microbiol. 2009;73:367–381. doi: 10.1111/j.1365-2958.2009.06775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berry J.D., Rajaure M., Young R. Spanin function requires subunit homodimerization through intermolecular disulfide bonds. Mol. Microbiol. 2013;88:35–47. doi: 10.1111/mmi.12167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jackson M. The mycobacterial cell envelope-lipids. Cold Spring Harb. Perspect. Med. 2014;4:a021105. doi: 10.1101/cshperspect.a021105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Summer E.J., Liu M., Gill J.J., Grant M., Chan-Cortes T.N., Ferguson L., Janes C., Lange K., Bertoli M., Moore C., et al. Genomic and functional analyses of Rhodococcus equi phages ReqiPepy6, ReqiPoco6, ReqiPine5, and ReqiDocB7. Appl. Environ. Microbiol. 2011;77:669–683. doi: 10.1128/AEM.01952-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pope W.H., Mavrich T.N., Garlena R.A., Guerrero-Bustamante C.A., Jacobs-Sera D., Montgomery M.T., Russell D.A., Warner M.H., Science Education Alliance-Phage Hunters Advancing Genomics and Evolutionary Science (SEA-PHAGES) Hatfull G.F. Bacteriophages of Gordonia spp. display a spectrum of diversity and genetic relationships. MBio. 2017;8:e01069-17. doi: 10.1128/mBio.01069-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.