Fig. 6.
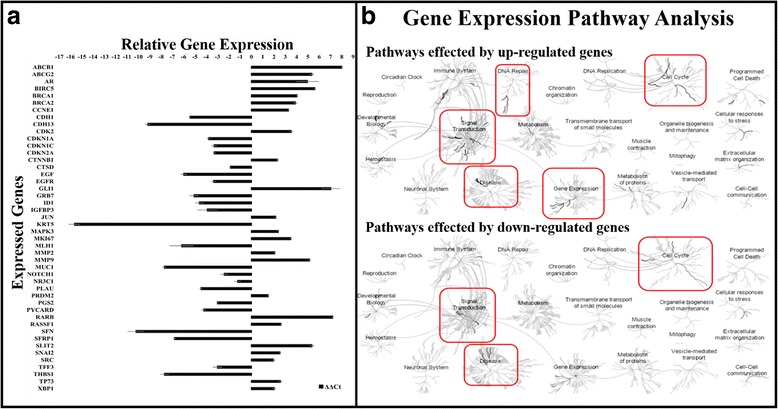
a Gene expression analysis of breast cancer KAIMRC1 cells utilizing breast cancer gene panel. 84 genes were analyzed by qPCR for gene expression changes out of which 46 genes showed significant changes. Data is plotted as relative up-regulation or down-regulation over normal breast MCF10A cells. Each column represents a single gene and represents data from duplicates. X-axis = Genes and y-axis = Relative gene expression (ΔΔCt) to normal breast cells. b Gene expression analysis of KAIMRC1 cells using bioinformatics pathway browser tool (Reactome). We segregated the identified genes into up- and down-regulated genes. Web-based freely available pathway analysis tool, Reactome Pathway database (reactome.org) was used to identify the pathways affected by these genes. In KAIMRC1 cells, upregulated genes were found to be activating DNA repair, signal transduction, metabolism of proteins and cell cycle related pathways whereas downregulated genes were mainly involved in signal transduction, cell cycle and disease related pathways
