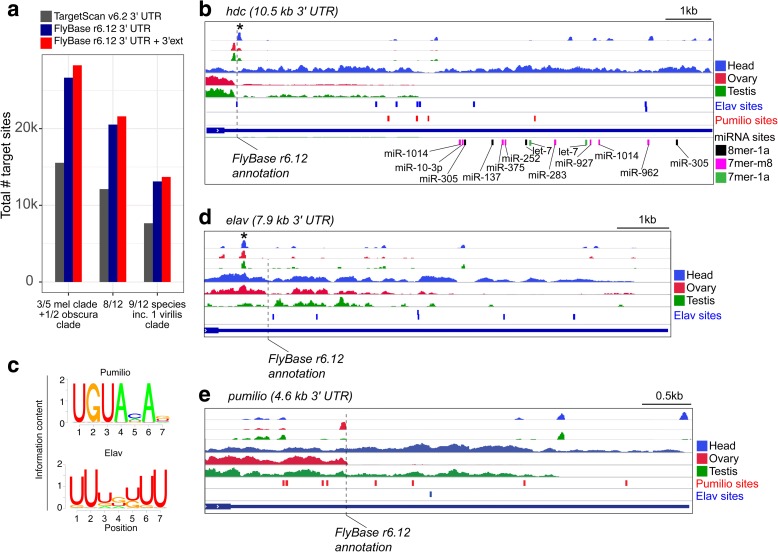
Fig. 5.
Annotation of well-conserved miRNA, Elav, and Pumilio binding sites. a We used TargetScanS algorithm to perform target predictions using the 3′ UTR databases used in the current available releases of TargetScanFly (v6.2), FlyBase (r6.12), and our current 3′ UTR annotations based on 3′-seq atlas (FlyBase + ext). This graph reports only 7mer-m8 seed matches to pan-Drosophilid conserved miRNAs, bearing conservation properties across the indicated fly species. b Example of headcase (hdc). TargetScanFly (v6.2) annotates no miRNA binding sites in the short 3′ UTR; however, an abundance of deeply conserved miRNA, Elav, and Pumilio binding sites exists in the much longer 3′ UTR that is well-expressed in head. c Position weight matrices for Pumilio and Elav binding. d The elav 3′ UTR has an abundance of conserved Elav binding sites. e The pumilio 3′ UTR has an abundance of conserved Pumilio binding sites. RNA-seq and 3′-seq are shown for each gene example. Asterisks denote likely false positive 3′-seq signal deriving from internal priming of genomically templated A-rich sequence