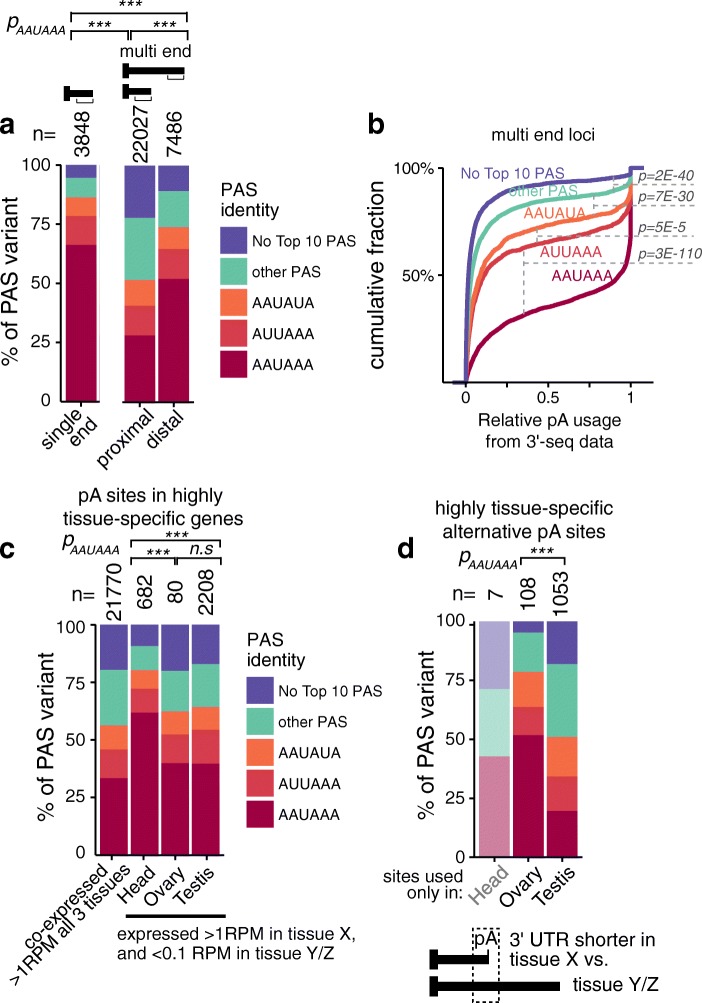
Fig. 6.
Distinctive properties of alternative and tissue-specific poly(A) sites. a Percentages of PAS types present in the 50-nucleotide window upstream of the pA site. pA sites were divided based on 3′ UTR class and position. Single end, pA sites that define the 3′ end of genes with only evidence of single terminal 3′ UTR isoform; proximal, pA sites found in the 3′ UTR of genes that undergo APA, all pA sites upstream of the most distal one; distal, the last pA sites of terminal 3′ UTRs that undergo APA. Proximal sites of APA genes exhibit distinctively lower fraction of optimal PAS. b Empirical cumulative distribution functions (ECDF) of the relative strength of all pA sites in terminal 3′ UTRs subdivided by the identity of the PAS (as shown in the legend on the left) in the 50 nucleotides upstream of the cleavage site. The relative strength score represents the fraction of isoforms ending at a specific site compared to downstream sites. This was calculated from the fraction of reads at each pA out of all reads at that site or downstream ones. The relative pA site usage of each PAS variant class compartmentalizes well according to their known efficiency of cleavage. The significant differences of the relative strength score among PAS identities were calculated by Wilcoxon rank-sum tests. c Analysis of PAS identity within tissue-specific genes. We compared genes that were co-expressed in the three tissues (> 1 RPM in head, ovary, and testis) vs ones that were deemed tissue-specific (> 1 RPM in one of these tissues, < 0.1 RPM in the other two). All PAS in these tissue-specific genes were plotted. The head utilizes a distinctively elevated fraction of optimal PAS. d Analysis of PAS identity at tissue-dominant pA sites. Percentages of PAS types that present in the 50-nucleotide window upstream of the pA site. pAs were divided based on tissue dominance in either head (n = 7), ovary (n = 108), or testis (n = 1053). Data for the head are opaque since so few available sites are head-specific. Testis APA sites utilize a broader range of suboptimal PAS compared to ovary APA sites. *** p < 0.0005; n.s. p > 0.05