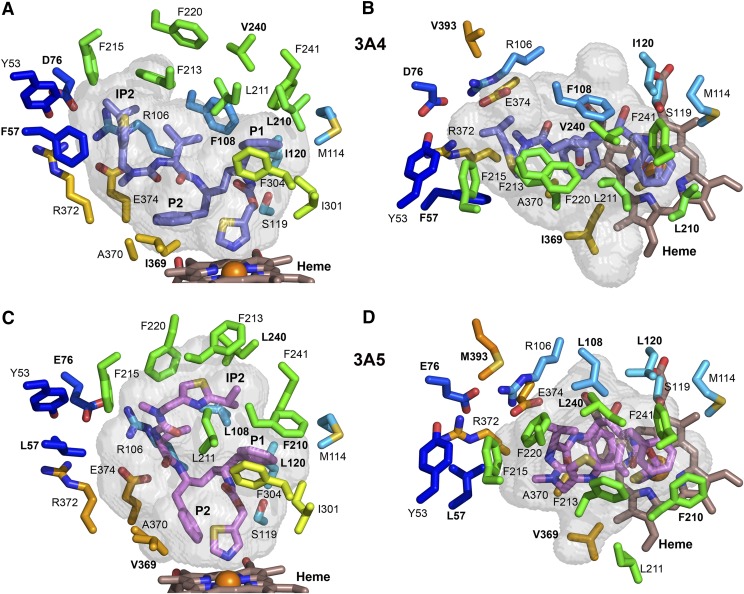
Fig. 4.
Differences in the identity and positioning of selected, conserved residues that shape the substrate binding cavities of chain A of 3A4 (A and B) and of 3A5 (C and D). Amino acids that differ between 3A4 and 3A5 are labeled with bold text. The longer F210 pushes the protein chain of 3A5 outward and displaces residue 211 away from the P1 group of ritonavir, which enlarges the upper portion of the cavity relative to 3A4. Additionally, the shorter L108 together with outward positioning of L108 Cα contribute to the larger upper cavity of 3A5. P2 of ritonavir displaces residues 369 and 370 on β1-strand 4 outward in 3A4. The smaller V369 does not contact the thiazole group of ritonavir or P2 in 3A5, which adopts a different orientation owing to the upward extension of ritonavir. The color scheme for carbons and heteroatoms is defined in earlier figure legends, and the views correspond to those in Fig. 3.