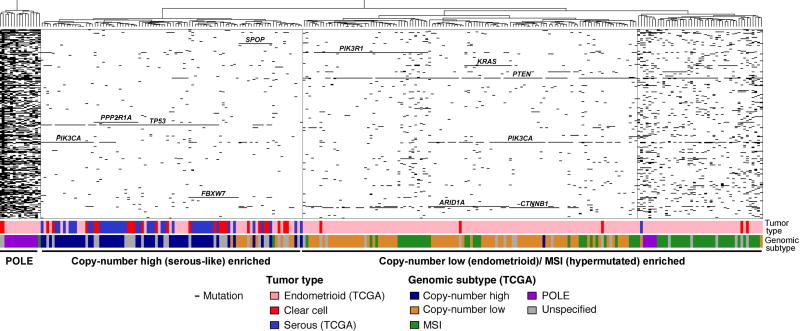
Figure 4. Hierarchical clustering of endometrial clear cell carcinomas from this study and endometrioid and serous carcinomas from TCGA using somatic mutations identified in 300 cancer genes.
Hierarchical cluster analysis of mutations identified in the 300 cancer genes included in our targeted massively parallel sequencing assay using Euclidean distance metric and Ward’s algorithm, including all endometrial clear cell carcinomas (ECCs) from the current study, and all endometrial endometrioid and serous carcinomas from TCGA. Three stable clusters were identified (supplementary material, Figure S5): a POLE cluster, a cluster enriched for endometrial carcinomas of copy-number high (serous-like) subtype, and a cluster enriched for endometrial cancers of copy-number low (endometrioid) and of MSI (hypermutated) subtypes. The tumor type as well as the molecular subtype of the endometrial endometrioid and serous carcinomas as defined by TCGA are presented in the phenobar below the heatmap, color-coded according to the legend. The majority of ECCs clustered with the serous carcinomas/ copy-number high (serous-like) tumors, however ECCs were also found in the POLE and the copy-number low (endometrioid)/ MSI (hypermutated) enriched clusters.