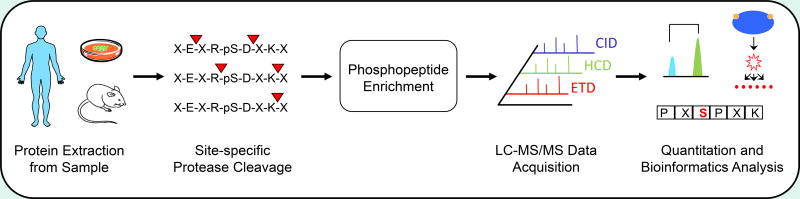
Figure 1.
Typical bottom-up phosphoproteomics workflow. Proteins are first extracted from cell lines, tissues, or biofluids and digested into peptides by site-specific proteases such as GluC, trypsin, or LysC. Phosphopeptides are then enriched and subjected to tandem mass spectrometry before bioinformatics analyses, including quantitation, signal transduction pathway mapping, or kinase motif extraction.