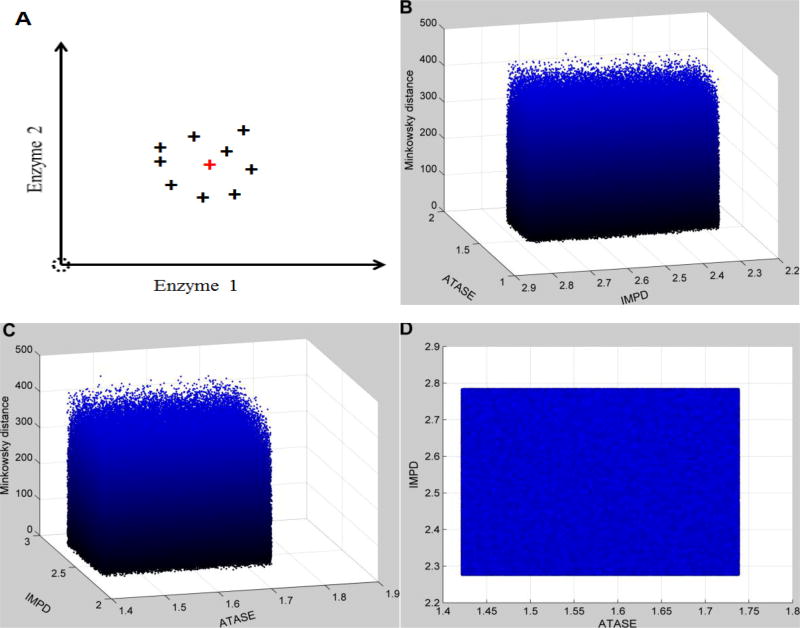
Figure 3. Distance topology for the Neighborhood surrounding the targeted enzymatic changes using absolute changes.
For human renal cell carcinoma, the observed enzymatic changes include the activation of IMPD (2.53 in fold change) and ATASE (1.58 in fold change). This targeted set of enzymatic changes is disturbed by uncertainty (10% relative noise sampled from a normal distribution). In addition, all other enzymes are also affected by 10% relative noise over normal activities. The x- and y-axes represent relative enzymatic activities of IMPD and ATASE in regard to their normal values, respectively. The z-axis shows the Minkowski distances between the simulated metabolic profiles and the targeted disease profile, using absolute changes. Subplot A schematically illustrates the uncertainty surrounding the targeted enzymatic changes (red cross). The same distance topology is viewed from different angles. B: horizontal rotation (−105) and vertical elevation (20); C: horizontal rotation (−15) and vertical elevation (20); D: horizontal rotation (0) and vertical elevation (20). Distances are similarly distributed within the neighborhood surrounding the targeted enzymatic changes.