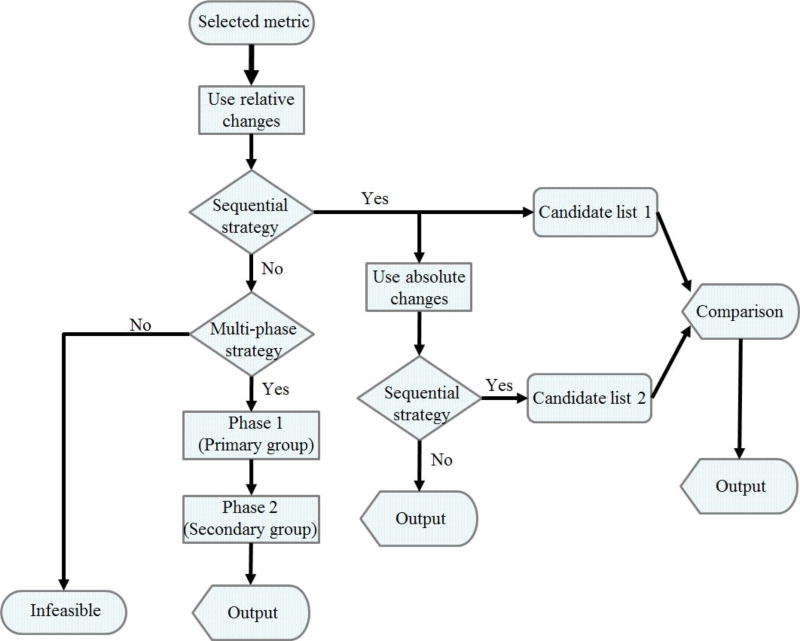
Figure 7. Strategic guidelines for the algorithmic inference of biochemical mechanisms from metabolomics data.
The flow chart shows recommendations for designing an algorithm for the inference of biochemical mechanisms underlying a disease from metabolomics data. Preferred metrics are Minkowski distance, Euclidean distance, Manhattan distance, Jeffreys & Matusita distance, Dice’s coefficient, and Jaccard similarity coefficient. These metrics have similar performance. The outputs from the sequential strategy and multi-phase strategy can be compared and provide further targets for experimental investigations.