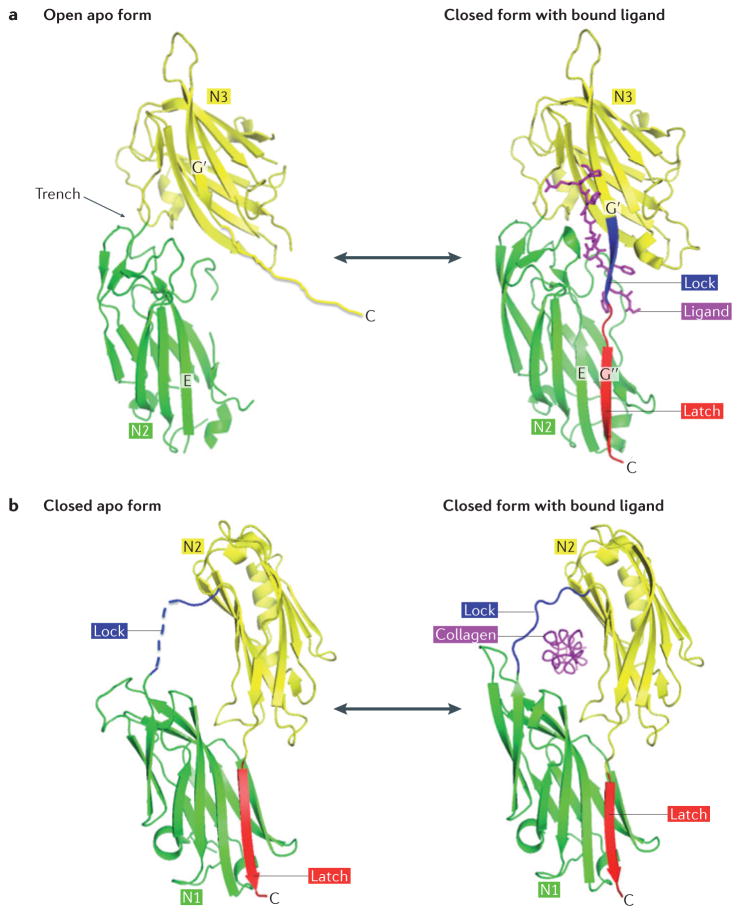
Figure 3. Mechanisms of ligand binding by MSCRAMM proteins.
a | Dock, lock and latch (DLL) mechanism. The A region of the microbial surface component recognizing adhesive matrix molecule (MSCRAMM) protein in its open apo form has a wide trench between the N2 and N3 subdomains (apo serine–aspartate repeat-containing protein (SdrG); Protein Data Bank (PDB) reference: 1R19). The ligand peptide (purple) inserts into this trench and the MSCRAMM protein undergoes conformational changes to a closed form and locks the ligand in place (SdrG–ligand complex; PDB reference: 1R17). In the apo form, the disordered carboxy-terminal extension of the N3 subdomain is not part of the crystal structure. After ligand binding, this region forms the lock (blue) and the latch (red), thus locking the ligand in place. b | Collagen hug mechanism. In the apo form, collagen adhesin (Cna) is in an equilibrium between an open and closed form. The crystal structure shows the closed form with the empty ligand-binding trench covered by the lock (blue) (apo Cna; PDB reference: 2F68). The latch peptide (red) has undergone β-strand complementation with the latching sequence in N1. In the open form (not shown), the rope-like collagen triple helix docks into a trench that is located between the N1 and N2 subdomains. The conformational change back to the closed form captures (or ‘hugs’) the ligand (purple) using residues in the lock region (blue) (Cna–collagen complex; PDB reference: 2F6A).