FIGURE 2.
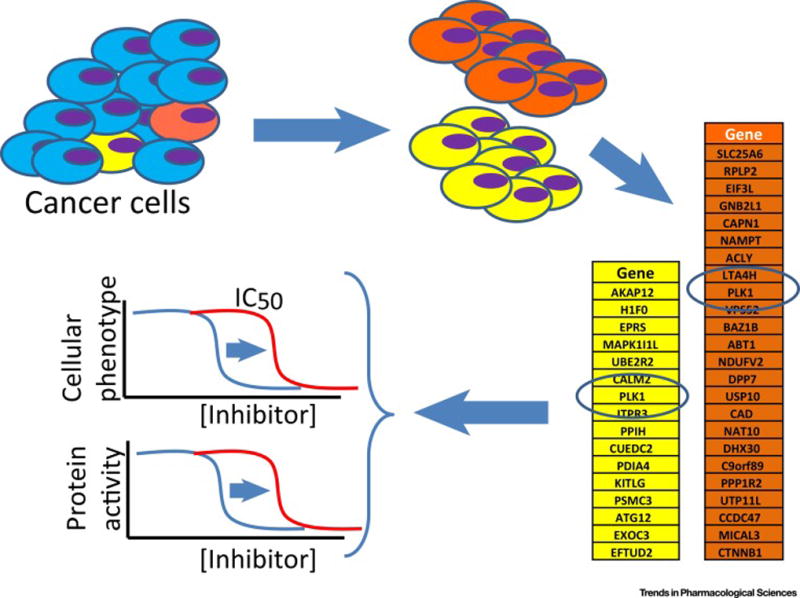
Schematic for DrugTargetSeqR. A genetically heterogeneous population of HCT116 cells is treated with the chemical inhibitor of interest. Clones that have reduced sensitivity to the inhibitor are isolated and expanded separately. Clones resistant to multidrug resistance (MDR) substrates are excluded from further analyses. Transcriptomes of six to eight clones and the parental cells are sequenced. Genetic differences between each clone and the parental cell population are identified. Genes that are altered (e.g. mutated) in multiple independent clones are selected. These mutations are introduced in other drug sensitive cell lines and those mutations sufficient to confer resistance are considered to be in the gene likely encoding the physiological target of the inhibitor. Biochemical assays are then used to test inhibition of the target protein’s activity or direct inhibitor binding. ‘Gold standard’ proof of target is established when the same mutation reduces the chemical inhibitor sensitivity in both cell-based and biochemical assays.
