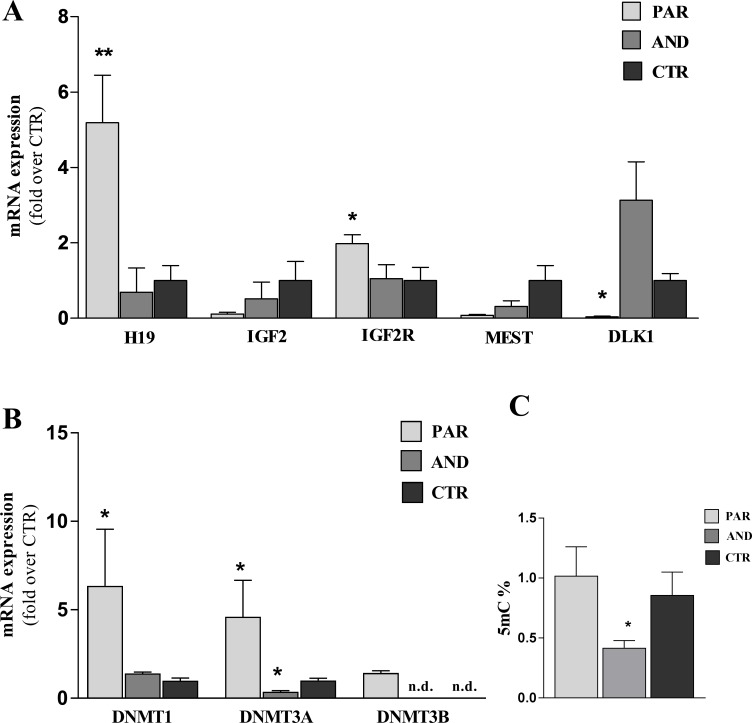
Fig 1. Epigenetic defects in uniparental placentae (chorion-allantoid tissues) 20 of pregnancy.
A) Maternally expressed genes, H19 and IGF2R, were overexpressed in PAR vs. CTR, while the paternally expressed gene DLK1 was down regulated in PAR vs. CTR (* indicates P<0.05; ** indicates P<0.01). No significant differences were detected for IGF2 and MEST. The pattern of expression was not “on” or “off” as expected, probably due to the perturbation of the epigenetic programming as a consequence of in vitro culture and abnormalities of our model. B) DNMT1, DNMT3A and DNMT3B were overexpressed in PAR vs CTR and DNMT3A was down expressed in AND vs CTR (* indicates P < 0.05, n.d. = not detected) C). Genome-wide methylation, assessed by 5-methylcytosine quantification, showed a significant hypomethylation in AND placentae compared to both PAR and CTR (* indicates P = 0.023).