Abstract
Troxerutin, a semi-synthetic derivative of the natural bioflavanoid rutin, has been reported to possess many beneficial effects in human bodies, such as vasoprotection, immune support, anti-inflammation and anti-aging. However, the effects of troxerutin on genome-wide transcription in blood cells are still unknown. In order to find out effects of troxerutin on gene transcription, a high-throughput RNA sequencing was employed to analysis differential gene expression in blood cells consisting of leucocytes, erythrocytes and platelets isolated from the mice received subcutaneous injection of troxerutin. Transcriptome analysis demonstrated that the expression of only fifteen genes was significantly changed by the treatment with troxerutin, among which 5 genes were up-regulated and 10 genes were down-regulated. Bioinformatic analysis of the fifteen differentially expressed genes was made by utilizing the Gene Ontology (GO), and the differential expression induced by troxerutin was further evaluated by real-time quantitative PCR (Q-PCR).
Introduction
Troxerutin (TX), known as vitamin P4, is a semi-synthetic derivative of the natural bioflavonoid rutin (alias rutoside, vitamin P) which is originally extracted from Ruta graveolens L. It is abundant in capers, olive, buckwheat, asparagus, and raspberries[1–4]. Compared with rutin, TX can be absorbed more easily through digestion system. The safety of TX has been confirmed in human including pregnant women and the aged[5–7], clinical trials have proved that TX has a very good safety profile and tolerability even at high doses[8].
Accumulative evidence has shown that TX has broad pharmacological effects, such as vasoprotection, anti-inflammation, immune support and anti-aging[9–16]. It is clinically recognized as an effective agent in treatment of cardiovascular diseases, for instance, varicose veins and chronic venous deficiency[17,18]. It has been reported that TX can pass through the blood-brain barrier and influence the nervous system[19,20]. So far, the effects of TX on genome-wide expression have not been reported, which could help understanding the clinical effects of TX.
In molecular biology, transcript identification and quantification of gene expression has been distinct core activities[21]. The rapid development of high-throughput sequencing of mRNA (RNA-Seq), for example, Illumina RNA sequencing, has provided efficient technology for transcriptomic characterization in human and other well-documented experimental animals like mice[22]. Theoretically, RNA-Seq can characterize every aspect of transcriptional activities. In this context, we used Illumina HiSeq 4000 to generate a substantial dataset of transcript reads in different groups treated with or without TX. To understand the transcriptomic effects of TX, bioinformatics analysis was made which contains Gene Ontology (GO) analyses, and real-time quantitative PCR (Q-PCR) was used to notarize the differential expression of genes induced by TX.
Materials and methods
Chemicals
TX was purchased from the Abcam Chemical Company (USA). RNAiso Blood RNA Extraction kit was purchased from Takara, PrimeScriptTM RT reagent kit with gDNA Eraser and real-time quantitative PCR kit were also obtained from Takara Bio (Dalian, China). Globin-Zero Gold rRNA Removal Kit for removal of unwanted ribosomal RNA and hemoglobin mRNA from mammalian blood RNA samples were bought from Illumina, Inc. (USA).
Animal care and maintenance
Male Kunming mice (Swiss mice, SPF), five-weeks old, were purchased from the Experimental Animal Center of Hainan province. They were housed at temperature of 22 to 24 ºC with 12-h light/12-h dark cycle. The mice had free access to water and normal chow diets until they were sacrificed with CO2. All efforts were made to alleviate suffering. All animal procedures were abide by the Manipulative Technique for the Care and Use of Laboratory Animals (2nd revision) issued by the State Scientific and Technological Commission of China. All animal experimental protocols were approved by the Institutional Animal Care and Use Committee of Hainan University (Hainan, China). All animals were randomly divided into control group and experimental groups (N = 3). The control group received injection of normal saline (NS), and the experimental group received subcutaneous injection of TX dissolved in NS at dose of 130 mg/kg twice daily at 9:00 AM and 5:00 PM. On the 3th day of treatment, the blood samples were drawn from the animals’ hearts 4 h after the last dose of TX.
RNA isolation and manipulation
Total RNA was extracted from the blood samples by using a RNA Extraction kit (Takara RNAiso blood). The concentration of RNA in each sample was determined using a NanoDrop 2000 micro-volume spectrophotometer (Thermo Scientific, USA), and the quality of RNA samples were examined using an Agilent Bioanalyzer (Agilent Technologies, USA). The defined criterion for qualification of RNAseq is that the RIN value of a sample is higher than 8.5. Ribosomal RNA and hemoglobin mRNA from the blood RNA samples was removed using a Globin-Zero Gold rRNA Removal Kit (Illumina, USA).
Sequencing library construction and illumina sequencing for transcriptomic analysis
The sequencing library construction and Illumina sequencing were performed by AnnoRoad Biotechnology Co, Ltd, China. Briefly, mRNA was enriched via purification with oligo (dT) magnetic beads and fragmented into short fragments using fragmentation buffer. First-strand cDNA was synthesized using random primers and fragments of mRNA as templates. Subsequently, second-strand cDNA was synthesized using DNA polymerase I, buffer, dNTPs, and RNaseH. After purification, we used a QIAQuick PCR kit and eluted by EB buffer. Purified double-stranded cDNA were end-repaired, added bases A, and ligated with sequencing adaptors. After SDS-PAGE, amplified products were gel extracted. The cDNA were PCR-amplified for a minimum number of cycles to avoid normalization for downstream quantitative gene expression analyses. Then it was sequenced on an Illumina HiSeq 4000 with the sequencing strategy PE150.
Reads processing and identification of differentially expressed genes
To ensure reproducibility and reliability of the results, quality control checks were applied pertinently at different stages of the analysis. Quality control for the raw reads contained the analysis of sequence quality, GC content, the presence of adaptors and low-quality reads. We adopted DESeq to analyze differential expression of genes (DEGs). Compared with the reference genome of Kunming mice, we selected standard was |log2Ratio|≥1 and q<0.05, then got up-regulated and down-regulated genes. All expressed genes were monitored, and their gene functions were explored using database annotations such as NR, NT, UNIPROT, GO, and KEGG.
Real-time quantitative PCR
Reverse transcription was carried out using PrimeScriptTM RT reagent kit with gDNA eraser (Takara, China). Total RNA was isolated from the blood samples drawn from mice hearts. For cDNA synthesis, one μg of total RNA was reverse-transcribed in a total volume of 20 μL, using a One-step gDNA Removal and cDNA Synthesis Super Mix kit (Trans Script).
Synthesized cDNA samples were diluted 3 times prior to Q-PCR. The primers were designed for 9 genes (Table 1). Q-PCR was accomplished using the SYBR premix Ex Taq kit (Takara, China). β-actin was used as the internal reference gene in this study. The results were analyzed on an ABI StepOnePlus Real-Time PCR System (Thermofisher Scientific, USA).
Table 1. Q-PCR primers.
| Target gene | Primer sequence(5’ to 3’) | Product length (bp) | |
|---|---|---|---|
| β-actin | Forward | GGCTGTATTCCCCTCCATCG | 154 |
| Reverse | CCAGTTGGTAACAATGCCATGT | ||
| Tnr | Forward | GGAGGTGACTACAGAAAGGGC | 137 |
| Reverse | AGAGGCTTTCAAGTGGCACG | ||
| Robo1 | Forward | TCCAAAGAGAACTGGGGAATGT | 142 |
| Reverse | GCTCCAGATGGGCGGTAG | ||
| Pnp2 | Forward | CGACCTCAAGTGGCAGTGAT | 148 |
| Reverse | GCAATCCAAACACCAGTCGG | ||
| H2-k1 | Forward | TGGACGACACGGAGTTCG | 147 |
| Reverse | CCACTCGGAAACTCTGCTCAT | ||
| Fcnb | Forward | CTGACTGTCCATGCGGCTG | 147 |
| Reverse | TCCTCTATCTCCTTTGGCACC | ||
| Arhgdig | Forward | GCTTGGTCAAGTACAAGCAGG | 126 |
| Reverse | CCATGATGATAGGCCCTGGAG | ||
| Hbq1a | Forward | CGGAATCTACACGACCGAGG | 146 |
| Reverse | TAGCGAGAGTCAGTGCATCG | ||
| Srp14 | Forward | CGTGTTCATCACCCTCAAGAAAT | 136 |
| Reverse | CACGGTGCTGATCTTCCTTTT | ||
| Npy | Forward | GGCCAGATACTACTCCGCTC | 135 |
| Reverse | CTTGTTCTGGGGGCGTTTTC | ||
Q-PCR was performed with 2 μl of template cDNA, 0.8 μl of forward primer (10 μM), 0.8 μl of reverse primer (10 μM), and 10 μl of SYBR Primer Ex Taq II (Tli RNaseH Plus) (2 X) in a total reaction volume of 20 μl. The procedure was conducted as follows—95°C for 30 s for initial denaturation followed by 40 cycles of 95°C for 5 s, and 60°C for 34 s—and then generated the melt curves for verification of amplification specificity by a thermal denaturing step. Genes was normalized to β-actin expression and calculated using the equation: change (x-fold) = 2-ΔΔCt[23].
Experiment data were analyzed using IBM SPSS statistics 19 software. Statistical significance was determined by the Student’s t test or one-way analysis of variance (ANOVA), and the differences were considered statistically significant at a value of P < 0.05.
Results
Illumina sequencing and data quality
Initial llumina sequencing results existed in the original image data file. Using the CASAVA softwarh the base calling function, they were transformed into sequenced reads, which were called the raw data. The raw data were stored in FASTQ file format. FASTQ files include the name of every read, base sequence and their sequencing quality information.
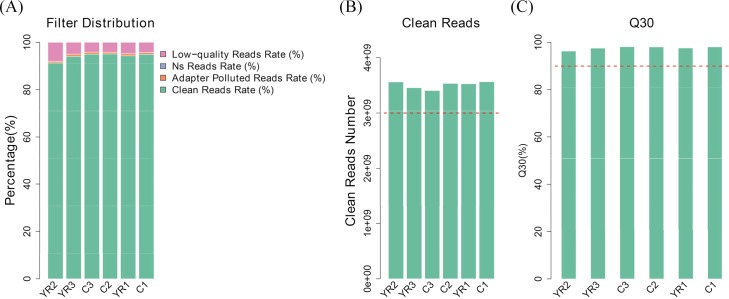
The raw reads output from the Illumina sequencing platform were trimmed for the purpose of quality control. Adaptor, N< 10% and low-quality reads were removed to obtain high-quality reads, which was called clean reads, producing about 23.76 million paired-end reads per sample equivalent to over 3.405 to 3.564 billion nucleotides per sample. Among those reads, over 96.27% of the clean reads with high-quality scores at the Q30 level (a base quality greater than 30 and an error probability of 0.1%) were identified (Table 2).
Table 2. Summary of statistical data for the transcriptome of mice.
| Samples | C1 | C2 | C3 | YR1 | YR2 | YR3 |
|---|---|---|---|---|---|---|
| Total raw reads | 25,037,206 | 24,771,616 | 23,903,170 | 24,938,022 | 25,995,740 | 24,499,022 |
| Total clean reads | 23,764,936 | 23,558,544 | 22,702,076 | 23,519,160 | 23,737,328 | 23,042,794 |
| Clean bases Number | 3,564,740,400 | 3,533,781,600 | 3,405,311,400 | 3,527,874,000 | 3,560,599,200 | 3,456,419,100 |
| Clean reads rate (%) | 94.92 | 95.1 | 94.98 | 94.31 | 91.31 | 94.06 |
| Clean Q30 bases rate (%) | 98.04 | 98.03 | 98.11 | 97.58 | 96.27 | 97.52 |
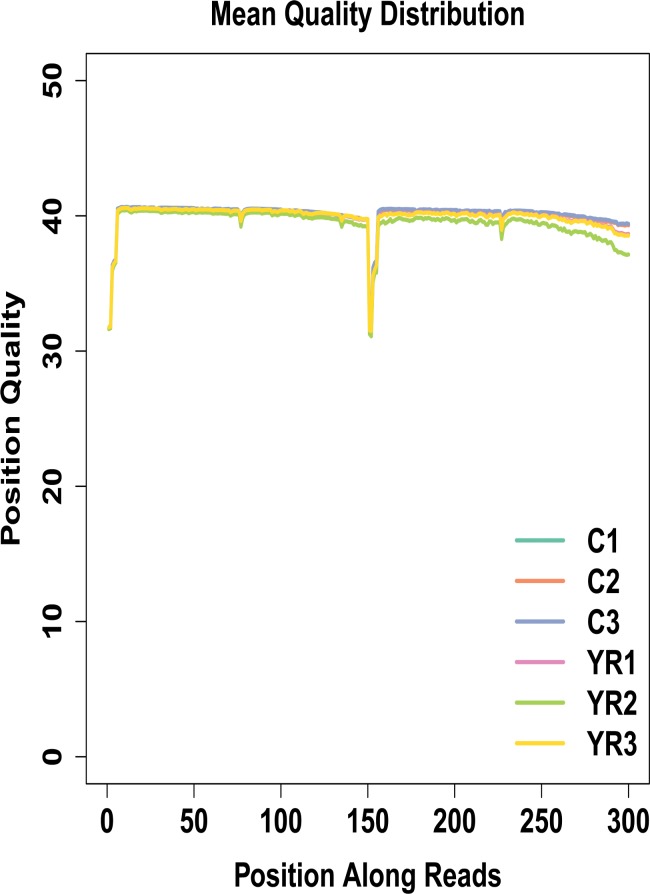
The clean reads were used for further analysis (Fig 1A and 1B). The cDNA libraries produced a total of 140,324,838 clean reads, which represented the majority of the data (almost covered 90% of the whole gene set), with Q30 scores > 95% (Fig 1C). The mean quality distribution of all samples was shown in Fig 2.
Fig 1.
A. Distribution of reads proportion before filtering. C1, C2 and C3 represent control groups that received injection of normal saline (NS), and YR1, YR2 and YR3 represent experimental groups that received subcutaneous injection of TX dissolved in NS at a dose of 130 mg/kg, twice daily. B. Data volumes of clean reads for samples. For the purpose of quality control, the raw reads output from the Illumina sequencing platform were filtered: adaptors, short reads with N < 10%, and low-quality reads were removed. C. Q30 proportion for samples. The clean reads with high-quality scores at Q30 levels (a base quality greater than 30 and an error probability less than 0.1%) are more than 95%. With a vast majority of bases scoring no less than the Q30 level, the levels of accuracy are ideal for sequencing applications.
Fig 2. Mean quality distribution of samples.
C1, C2 and C3 represent control groups that received injection of normal saline (NS), and YR1, YR2 and YR3 represent experimental groups that received subcutaneous injection of TX dissolved in NS at a dose of 130 mg/kg, twice daily. The x-axis indicates base position of filtered high quality sequences. The y-axis indicates mean sequencing quality at each position.
Comparison analysis with a reference genome and gene mapping
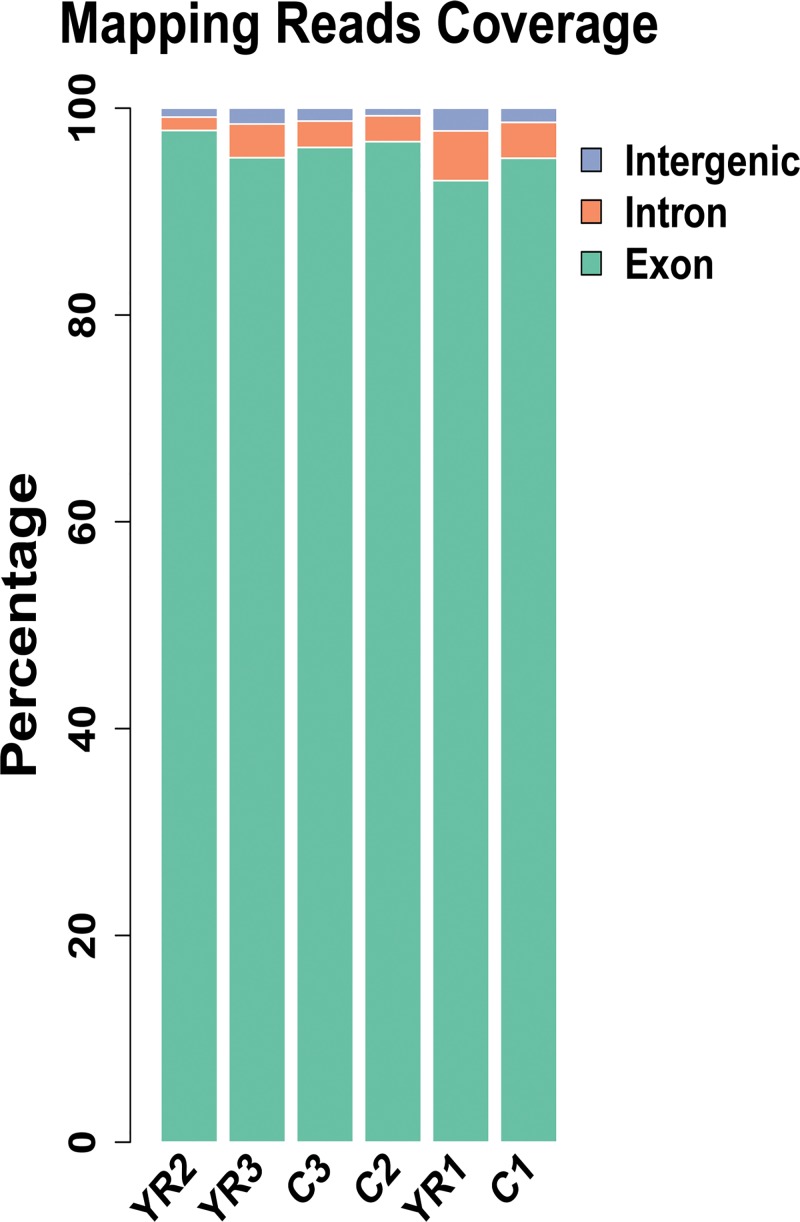
The filtered sequencing results were compared with the reference genome of Kunming mice for its orientation to the genome. For the samples, we used TopHat software, which was specifically designed for comparison of the transcriptome data[24,25]. To ensure accuracy and reliability of the data, about 140 million high-quality reads were generated, and over 88.5% of the data were mapped to the reference genome for each sample. Most of the sequences were compared to the exon region. Besides, alternative splicing and expression noise maybe come from the intron region, and the new transcripts and expression noise may belong to the intergenic sequence. For the control group and experiment group, 95.35% and 96.04% were mapping to the exon region, 3.12% and 2.84% belonged to the intron region, and 1.53% and 1.12% were in the intergenic sequence respectively (Fig 3) (Table 3).
Fig 3. Reads coverage mapping to the reference genome of Kunming (Swiss) mice.
C1, C2 and C3 represent control groups that received injection of normal saline (NS), and YR1, YR2 and YR3 represent experimental groups that received subcutaneous injection of TX dissolved in NS at a dose of 130 mg/kg, twice daily. Blue, green and orange refer to intergenic, exon and intron region respectively.
Table 3. Summary statistics of clean reads mapping.
| Sample | C1 | C2 | C3 | YR1 | YR2 | YR3 |
|---|---|---|---|---|---|---|
| Total Clean Reads | 23,764,936 | 23,558,544 | 22,702,076 | 23,519,160 | 23,737,328 | 23,042,794 |
| Mapping Reads | 22,158,309 | 22,118,461 | 21,098,768 | 20,836,785 | 21,276,557 | 20,227,093 |
| Mapped Rate (%) | 0.93 | 0.94 | 0.93 | 0.89 | 0.9 | 0.88 |
| Exon (%) | 95.16% | 96.76% | 96.20% | 92.99% | 97.84% | 95.21% |
| Intron (%) | 3.47% | 2.50% | 2.55% | 4.81% | 1.30% | 3.26% |
|
Intergenic (%) |
1.37% | 0.74% | 1.25% | 2.20% | 0.86% | 1.54% |
Analysis of differential expression of genes
Differential expression analysis requires that gene expression values be compared among samples. Reads Per Kilobase Million (RPKM) is used to quantitatively estimate value of gene expression[26], which has the computation formula:
Where R is the number of mappable reads in specific genes, N is the total number of reads mapped to genes in a particular sample, and L is the length of the gene exon.
In general, differential gene expression only account for a small portion of total gene. There is little effect on the distribution of the sample expression quantity by a few differentially expressed genes. All samples have similar overall gene expression pattern (S1 Fig).
Differences in gene expression in mice in two different groups were examined. For biological triplicate samples, we adopted DESeq to analyze differential expression of genes. In total, fifteen genes were differentially expressed, among which 5 genes were significantly up-regulated and 10 genes down-regulated in TX-treatment mice (S2 Fig). We identified their gene functions using database annotations tools such as NCBI, UNIPROT, and GO databases. We summarized 15 different genes main function influenced by TX in mice (Table 4).
Table 4. Summary of 15 genes for the transcriptome of mice.
| Gene | variation tendency | Main function |
|---|---|---|
| Tnr | up | cure neurological diseases and neuroprotection |
| Hbq1a | up | impact the activity of oxygen transporter, anti-oxidative |
| RPL17e | up | important for ribosome architecture and function, tumor suppressor |
| GM10499 | up | against extracellular microbes and toxic molecules |
| GM5526 | up | responsible for skeletal muscle tissue development and muscle filament sliding |
| Robo1 | Down | protect liver from injury and fibrosis |
| Pnp2 | Down | positively regulate T cell-mediated cytotoxicity and proliferation of T cells |
| H2-k1 | Down | anti-inflammatory and immune |
| Fcnb | Down | ameliorate inflammation |
| Arhgdig | Down | catalytic activity and protein localization |
| Srp14 | Down | protein export |
| Npy | Down | immunomodulatory function, maintenance of body homeostasis and inhibits diabetes |
| Rpl13e | Down | drug resistance, virus replication and anti-oxidative defense |
| GM28114 | Down | against extracellular microbes and toxic molecules |
| GM12366 | Down | anti-cancer |
Gene Ontology (GO) analysis for selected gene
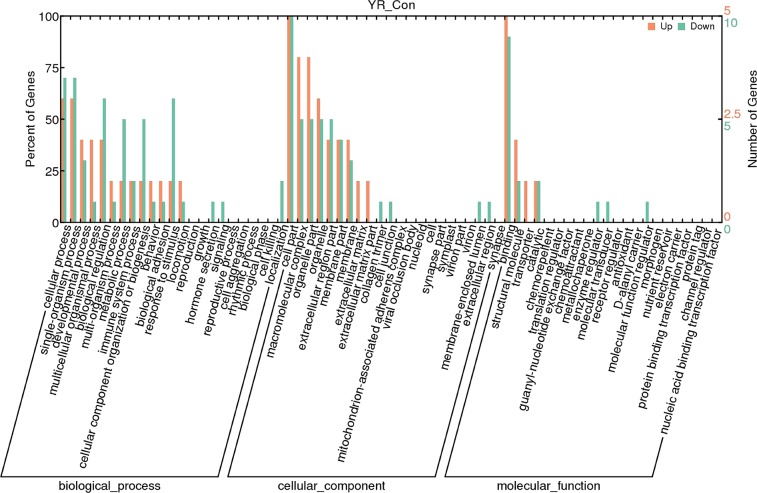
To further understand the function of the differential expression of genes, GO term enrichment analysis (q<0.05) was performed. There are three GO domains: biological process (GOBP), cellular component (GOCC), and molecular function (GOMF). The most significantly enriched GO terms were “single-organism process” and “biological regulation” within GOBP, ‘binding’ and ‘structural molecule’ were the two most abundant terms within GOMF, and “macromolecular complex” and “organelle part” were the most highly represented terms in GOCC respectively (Fig 4).
Fig 4. Gene Ontology (GO) classification of differentially expressed genes (DEGs).
DEGs are classified into three major domains: biological process (BP), cellular component (CC) and molecular function (MF). The left y-axis indicates the percentage of a specific category of genes in a domain. The right y-axis indicates the number of genes in the category. (Con, control group; YR, experimental group).
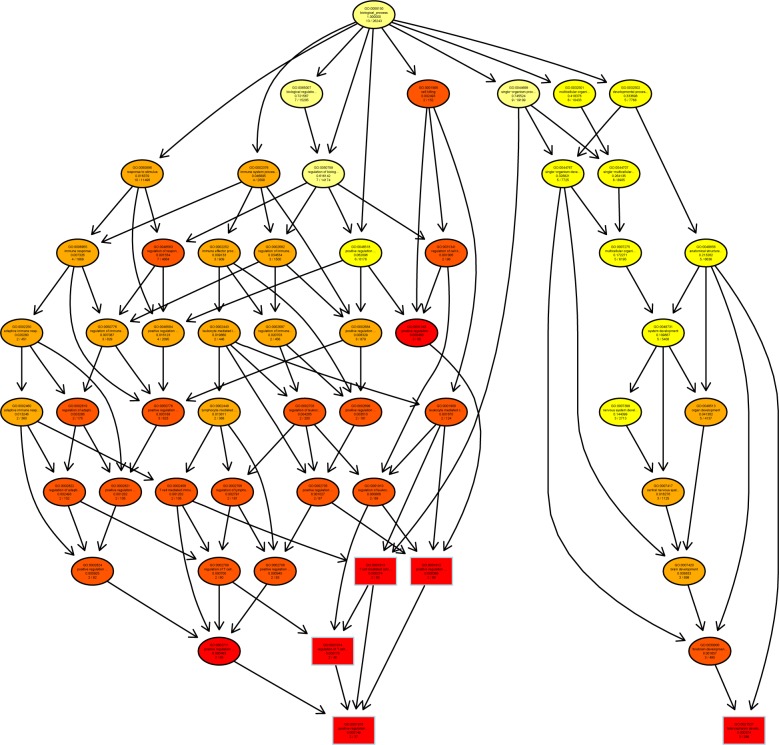
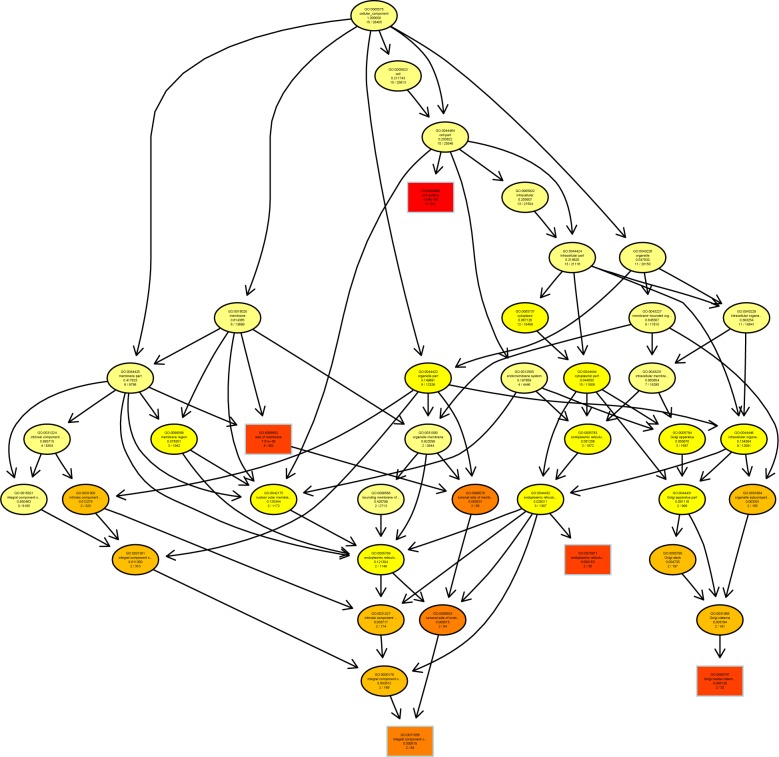
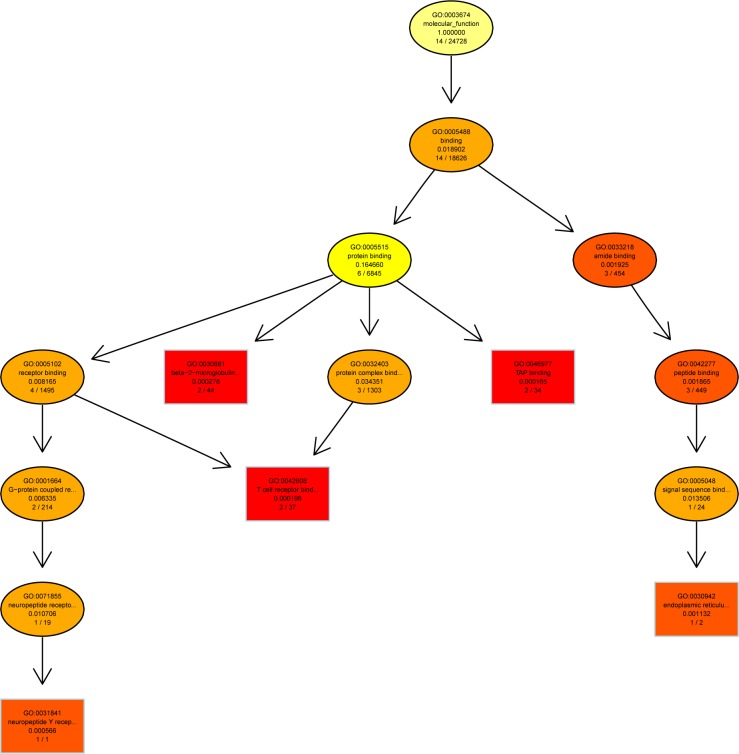
A directed acyclic graph (DAG) showed enrichment analysis results. It is a branch inclusion relationship that the lower node function was subordinate to the upper nodes function. Top 5 GO enrichment analysis results were selected as master nodes in DAG. The color depth of the DAG nodes indicates the degree of enrichment, the darker the red color is, the higher the enrichment degree of the node would be.
We found that genes involved in T cell mediated cytotoxicity (q = 0.000149) and telencephalon development (q = 0.000374) were most highly enriched (Fig 5). Further characterization of the differential expression of genes using DAG showed that the differentially expressed genes were implicated in cell surface, cell membrane (q = 5.04e-06) and endoplasmic reticulum exports (q = 0.000153) (Fig 6). In molecular function, genes in β2-mioroglobule binding (q = 0.000278), TAP-binding (q = 0.000165), T cell and neuropeptide Y receptor binding (q = 0.000196) were highly enriched (Fig 7).
Fig 5. Directed acyclic graph (DAG) for differentially expressed genes (DEGs) in the biological process (BP) ontology.
Fig 6. Directed acyclic graph (DAG) for differentially expressed genes (DEGs) in the cellular component (CC) ontology.
Fig 7. Directed acyclic graph (DAG) for differentially expressed genes (DEGs) in the molecular function (MF) ontology.
Real-time quantitative PCR
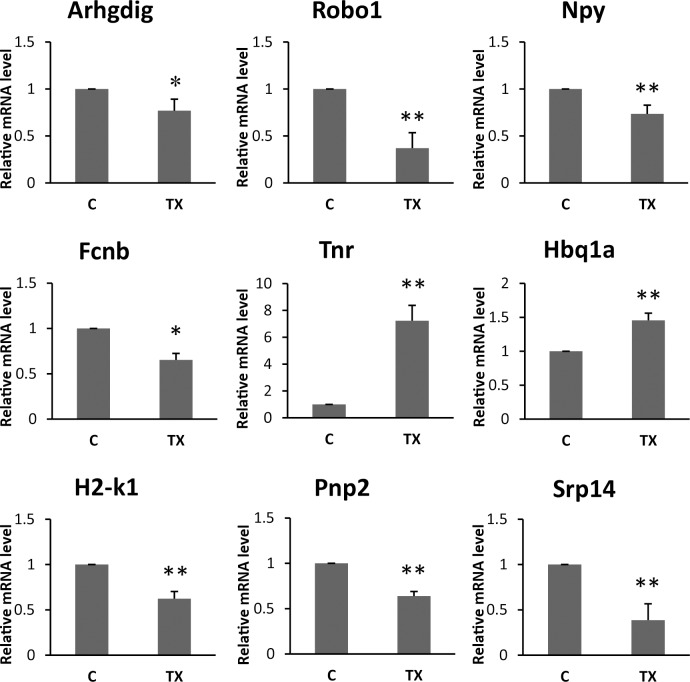
To further confirm the accuracy and reproducibility of the RNA-Seq results, real-time quantitative Q-PCR was performed to quantify the transcriptional levels. Nine differentially expressed genes, including Tnr, Robo1, Npy, Arhgdig, Srp14, H2-k1, Pnp2, and Fcnb, Hbq1a were evaluated, and each genes showed significantly differential expression after the treatment with TX (Table 5) (P<0.05, Fig 8). We performed the transcriptomic characterization of the effects of TX in mice to identify differentially expressed genes related with pharmacological properties of TX (Fig 9).
Table 5. Relative expression values determined using RNAseq and real-time quantitative PCR (Q-PCR).
| gene | RNA-seq | Q-PCR(±SD) | variation tendency | ||
|---|---|---|---|---|---|
| Fold Change | Log2Fold Change | Relative expression values(2−ΔΔCT) | RNA-seq | Q-PCR | |
| Tnr | 187.9821351 | 7.554451752 | 6.7992±1.27 | Up | Up |
| Robo1 | 0.005668298 | -7.462868797 | 0.3707±0.11 | Down | Down |
| Pnp2 | 0.024258529 | -5.365364134 | 0.5447±0.09 | Down | Down |
| H2-k1 | 0.127654812 | -2.969680168 | 0.7192±0.07 | Down | Down |
| Fcnb | 0.036825424 | -4.763154068 | 0.5041±0.04 | Down | Down |
| Arhgdig | 0.015302976 | -6.030043964 | 0.7694±0.12 | Down | Down |
| Hbq1a | 298.7735665 | 8.222908703 | 1.4009±0.15 | Up | Up |
| Srp14 | 0.199374235 | -2.326449115 | 0.4807±0.10 | Down | Down |
| Npy | 0.000831662 | -10.23171562 | 0.3677±0.01 | Down | Down |
Fig 8. Q-PCR for nine differentially expressed genes (DEGs).
Relative gene expression was normalized by comparison with the expression of β-actin, and calculated using the 2−ΔΔCT. (C, control group; TX, troxerutin, s.c. injected at a dose of 130 mg/kg, twice daily. *P < 0.05 and ** P<0.01 vs control group, N = 6–8).
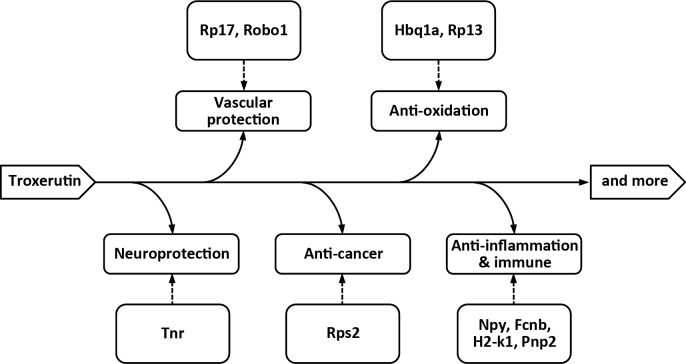
Fig 9. Pleiotropic effects of troxerutin.
Discussion
TX, a rutoside derivative, has raised considerable interest because of its extensive pharmacological activities. It has been conventionally used for treating diseases including capillary fragility, chronic venous and varicosity in clinical[27]. Researches over the past decades have revealed that TX can act againstγ-radiation-induced lipid peroxidation, UV radiation and colon carcinogenesis[28–30]. Furthermore, TX can cure retinopathy and play a significant role in the management of type-2 diabetes mellitus[31–33]. Beside, TX can inhibit 2-AA + UVA radiation-induced DNA damage and environmental carcinogens, showing its potential therapeutic activities for cancer[34]. Gene expression behind the pharmacological activities of TX has not been investigated clearly so far. And our findings provide novel insights into TX, which could help understanding clinical effects of TX.
In this study, we made a comprehensive analysis of the effects of TX on genome-wide expression of genes. A total of 15 differentially expressed genes (DEGs) were found, in which 5 genes were up-regulated and 10 genes down-regulated significantly. Moreover, Q-PCR confirmed that our RNA-Seq results were reliable. Our experimental findings provide evidence about the TX against diabetes, cardiovascular diseases and cancer through down- or up-regulation of different genes, implicating its treating capacity.
Tenascin-R (TN-R) is an extracellular matrix protein expressed primarily in the central nervous system. It is a member of the tenascin gene family, which includes 4 known members in vertebrates. During development of mammalian brain, TN-R is found around birth[35]. TN-R can affect cell migration, cell differentiation and adhesion. TN-R expression is tightly regulated in a spatiotemporal manner in brain, especially during cortical plate formation[36]. More importantly, many researches showed that TN-R could play an important role in brain development and cognition. By activating microglia, TN-R can indirectly elevate the secretion of cytokines and growth factors, including brain-derived neurotrophic factor (BDNF), transforming growth factor-β (TGF-β), CXCL2, and nerve growth factor (NGF)[37]. In addition, loss of TN-R was shown to impair cognition, synaptic plasticity and motor abilities[38]. And it has been proved to participate in maintaining a balance between excitatory and inhibitory circuits involved in learning, memory and cognition in mice[39–41]. TN-R deficient mice showed increased excitatory transmission through reduced perineuronal nets and altered synaptic activities[42]. In summary, TN-R may play a primary or secondary role in many neurological diseases [43] and have a significant role in neuroprotection. Previous evidence showed that TX significantly improved behavioral performance of D-galactose-treated mice in the step-through test and Morris water maze[44], and alleviated the oxidative damage caused by D-galactose in the liver and kidney, as well as cognitive impairments. In addition, some researchers think TX could be recommended as a possible candidate for prevention and therapy of cognitive deficits and Alzheimer's disease[45], and improved synaptic plasticity failure induced by Amyloid β peptide. The up-regulation of TN-R expression induced by TX may underlie its neuroprotective function.
Neuropeptide Y (NPY), a 36-amino acid peptide [46], is a highly conserved neurotransmitter involved in a broad range of fundamental physiological activities. NPY’s pleiotropic functions comprise regulation of body-weight and energy balance, brain activity, food intake, vasoconstriction, immune function, and emotional response[47]. In fact, its immunomodulatory function and maintenance of body homeostasis are also vital [48]. The effects of NPY are depend on many factors, such as at least four G protein-coupled receptors known as Y1, Y2, Y4, Y5 and cell types involved[49]. Different receptors are involved in various physiological activities. During inflammation, immune cells themselves are capable of producing and releasing NPY. NPY can regulate immune cell activities through a paracrine or autocrine mode of action[50–52]. NPY has different effects on inflammation. The binding of NPY to receptors influences the activities of immune cells in either a pro-or an anti-inflammatory manner[53]. Recently, it has been proved that NPY possesses a pro-inflammatory function in the gut[54,55]. Additionally, loss of NPY can combat obesity and diabetes through increased energy expenditure and lowered fat contents. In our study, we found that under the effect of TX, the NPY expression was suppressed. It provides evidence that TX can ameliorate lipid abnormalities and diabetes[56,57], and have anti-inflammatory activities at least in part.
Ribosomes are essential components of the protein synthesis machinery. Ribosomes are formed from two unequally sized subunits: the large ribosomal subunit and small ribosomal subunit[58]. The process of ribosome biogenesis is well organized and tightly regulated. There is increasing evidence indicating that ribosomal proteins (RPs) have extraribosomal functions including DNA repair and other cellular processes. It has been shown that RPs are related to the development and progression of metabolic, cancer and cardiovascular diseases [59]. 60S ribosomal protein L17 (rpl17) is a protein that is encoded by the Rpl17 gene. It has an essential role in ribosome architecture and function. Rpl17 is thought to stabilize long-range interactions important for establishing the structure of the polypeptide exit tunnel, during 60S subunit assembly[60]. Reducing the amount of rpl17 in mouse cells led to stalled 60S subunit maturation, causing degradation of most of synthesized precursors, which could affect normal functions of the ribosome. Elaine et al. discovered that rpl17 is a vascular smooth muscle cell (VSMC) growth inhibitor, akin to a tumor suppressor[61]. In our study, Rpl17 was found to be up-regulated by TX. It could have relationship with that TX cures cardiovascular diseases and is conducive to the formation of ribosomes. Besides, ribosomal protein L3 (rpl3) is known to be an essential component for the peptidyltransferase center[62]. It acts as a binding site for a ribosome inhibitory protein and plays an important role in drug resistance and virus replication[63]. Annapina et al. revealed that Rpl3 was down-regulated and the expression of stress-response genes was activated in rCalu-6 cells. They found that rpl3 reduced xCT and GST-a1 expression levels. Researches also demonstrated that rpl3 was a negative regulator of cystathionine-β-synthase (CBS) expression. Silencing CBS gene severely reduces cellular glutathione (GSH) levels, and down-regulation of Rpl3 can induce anti-oxidative defense. These represent indirect ways for TX to potentiate expression of antioxidant genes to activate defensive response. Rpl3 also regulates negatively the activation of NF-kB[64]. Rps2, a 32 kDa ribosomal protein, is over expressed in prostate cancer and promotes malignancy of human prostate PC-3ML cells in the severe combined immunodeficiency (SCID) tumor modeling studies[65]. Previous studies have found that Rps2 is overexpressed in human squamous cell carcinoma and breast tumor samples. Thus, rps2 might promote cancer and be an excellent therapeutic target for the treatment of the diseases. In this context, the effect of TX to inhibit RpS2 expression and thereby stabilize it in cells indicates that TX may have anti-cancer function.
The mammalian signal recognition particle (SRP) is an 11S cytoplasmic ribonucleoprotein that plays an essential role in protein sorting. It is a cytosolic particle that transiently binds to the endoplasmic reticulum (ER) signal sequence of a nascent protein, to the large ribosomal unit, and to the SRP receptor in the ER membrane[66]. Srp14 encodes SRP subunit 14 (SRP14). It can influence protein export, as specified in the Gene Ontology Molecular Function (GOMF). SRP14 and SRP9 proteins form a heterodimer that bind to the RNA of Alu domain of SRP which is responsible for translation arrest. This gene is significantly down-regulated by TX. Besides, GTP hydrolysis plays a vital role in the SRP cycle; SRP protein and both subunits of the SRP receptor contain G-domains. The GTPase cycle of SRP, modulated by the ribosome, provides the regulatory link between translation and translocation machineries[67,68]. Interestingly, in our study, Arhgdig was found to be down-regulation by TX. It’s Gene Ontology biological process (GOBP) is the regulation of catalytic activity and protein localization. This gene encodes a protein which is a GTPase activator. To sum up, our study indicated that that TX may have pharmacological effects on the synthesis and export of proteins.
Form the DAG, we found that the genes involved in T cell-mediated cytotoxicity and telencephalon development were highly enriched. T cells help eliminate pathogens present in infected cells and also help B cells make different kinds of antibodies to protect against extracellular microbes and toxic molecules[69]. To accomplish these important functions, T cells have to interact intimately with other cells and then come into play. However, T cells are unable to peek beneath the surface of cells to identify the cells that have ingested bacteria. T cells recognize antigenic peptide only in the context of MHC I or MHC II molecules that are displaying the antigen on cell surface[70]. MHC I molecules present peptides from the proteins that are synthesized by cells. This recognition allows the immune system to detect cells in abnormal states for elimination. Moreover, Reduction of MHC I molecule expression in human tumors is often detected[71,72]. Among DEGs, H2-k1 and GM10499 gene encode major histocompatibility complex class I (MHC-I), and GM28114 encodes lymphocyte antigen 6 complexes. Expression of these genes are differently affected by TX. In GOBP, they can positively regulate T cell-mediated cytotoxicity and endocytosis. Besides, PNP2 is a family of purine-nucleoside phosphorylase. It can also positively regulate T cell-mediated cytotoxicity and proliferation of T cells. PNP2 expression is down-regulated by TX.
Natural immunity depends on the ability of some pattern recognition molecules to sense molecular markers. Ficolin is a group of pattern recognition molecules to recognize molecules on pathogens, apoptotic and necrotic cells. A series of new findings have indicated that ficolin B encoded by Fcnb gene can activate the complement system via the lectin pathway similar to MBL[73–75]. The activation of the complement system results in the release of multiple inflammatory signaling molecules[76]. Recently, a number of reports have shown that membrane attack complexes (MACs) were produced by complement activation. MACs can increase production of pro-inflammatory cytokines IL-8 and NF-kB nuclear translocation. These cytokines and molecules can induce inflammation. Moreover, Ficolins are reported to stimulate expression of inflammatory cytokines by macrophages[77]. Under the effect of TX, the expression of Fcnb gene was down-regulated, which may ameliorate inflammation.
The Roundabout (Robo) family proteins are transmembrane receptors. Secreted proteins Slit-family proteins (SLITs) can act through Robo receptors to mediate axonal guidance and branching. It could be related with directing migration of many cell types, such as immune, and tumor cells[78]. Robo1 was initially found in Drosophila. SLIT/ROBO signaling is an important regulator of cellular interactions[79]. Some researchers have proved that Slit2-Robo1 signaling may debilitate liver injury and fibrosis[80]. In our study, Robo1 gene expression was decreased by TX, which could protect liver from injury and fibrosis. In addition, Lvzhen et al. demonstrated that silencing the expression of Robo1gene inhibited cell proliferation and suppressed the development of proliferative vitreoretinopathy (PVR), offering a potential therapeutic usefulness in treating PVR[81]. Other studies support that ovarian angiogenesis was enhanced by a partial lack of Robo1 genes and this lack would enhance ability of fertility[82].
In addition to the above, Hbq1a encodes hemoglobin, theta 1A, which is closely connected with oxygen and iron binding, and impact the activity of oxygen transporter. It may be associated with anti-oxidative effects of TX[83]. GM5526 is mainly responsible for skeletal muscle tissue development and muscle filament sliding. It encodes a protein, which is a structural constituent of muscle.
Conclusion
Our study accentuated the beneficial effects of bioflavonoid derivative TX by analyzing differential gene expression in blood cells. Transcriptomic analysis demonstrated that the expression of fifteen genes was significantly changed by the treatment with TX, among which 5 genes were up-regulated and 10 genes were down-regulated. DAG analysis showed that these genes could influence T cell-mediated cytotoxicity (q = 0.000149), telencephalon development (q = 0.000374), cell membrane (q = 5.04e-06), endoplasmic reticulum exports (q = 0.000153), β2-microglobule binding (q = 0.000278), TAP-binding (q = 0.000165), and T cell and neuropeptide Y receptor binding.
Among the 15 DEGs, TX has neuroprotective effects and improves cognitive impairment by up-regulating TN-R expression, anti-inflammatory effects and inhibits diabetes by down-regulating Npy and Fcnb, anti-cancer effects by changing ribosome protein gene rps2, effects on cardiovascular disease by up-regulating Rpl17 and down-regulating robo1, and anti-oxidative activities by changing Rpl3 and Hbq1a expression. In addition, H2-k1 and Pnp2 expression was also changed. They are involved in positive regulation of T cell-mediated cytotoxicity and T cell proliferation. Srp14 and Arhgdig expression were also change, which affect synthesis and export of proteins.
Supporting information
(PDF)
(PDF)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by a research fund from the National Natural Science Foundation of China (31460226, D.W.), and two research funds from the Natural Science Foundation of Hainan Province (317039, S.W. and 314058, D.W.).
References
- 1.Fan SH, Zhang ZF, Zheng YL, Lu J, Wu DM, Shan Q, et al. (2009) Troxerutin protects the mouse kidney from D-galactose-caused injury through anti-inflammation and anti-oxidation. International Immunopharmacology 9: 91–96. doi: 10.1016/j.intimp.2008.10.008 [DOI] [PubMed] [Google Scholar]
- 2.Sampath S, Karundevi B (2014) Effect of troxerutin on insulin signaling molecules in the gastrocnemius muscle of high fat and sucrose-induced type-2 diabetic adult male rat. Mol Cell Biochem 395: 11–27. doi: 10.1007/s11010-014-2107-2 [DOI] [PubMed] [Google Scholar]
- 3.Panat NA, Maurya DK, Ghaskadbi SS, Sandur SK (2016) Troxerutin, a plant flavonoid, protects cells against oxidative stress-induced cell death through radical scavenging mechanism. Food Chemistry 194: 32–45. doi: 10.1016/j.foodchem.2015.07.078 [DOI] [PubMed] [Google Scholar]
- 4.Panat NA, Maurya DK, Ghaskadbi SS, Sandur SK (2016) Troxerutin, a plant flavonoid, protects cells against oxidative stress-induced cell death through radical scavenging mechanism. Food Chem 194: 32–45. doi: 10.1016/j.foodchem.2015.07.078 [DOI] [PubMed] [Google Scholar]
- 5.Marhic C (1991) Clinical and rheological efficacy of troxerutin in obstetric gynecology. Rev Fr Gynecol Obstet 86: 209–212. [PubMed] [Google Scholar]
- 6.Munshi A, Hobbs M, Meyn R (2005) Clonogenic cell survival assay. Methods Mol Med 110: 21–28. doi: 10.1385/1-59259-869-2:021 [DOI] [PubMed] [Google Scholar]
- 7.Wijayanegara H, Mose J, Achmad L, Sobarna R, Permadi W (1992) A clinical trial of hydroxyethylrutosides in the treatment of haemorrhoids of pregnancy. J Int Med Res 20: 54–60. doi: 10.1177/030006059202000107 [DOI] [PubMed] [Google Scholar]
- 8.Maurya DK, Balakrishnan S, Salvi VP, Nair CK (2005) Protection of cellular DNA from gamma-radiation-induced damages and enhancement in DNA repair by troxerutin. Mol Cell Biochem 280: 57–68. doi: 10.1007/s11010-005-8052-3 [DOI] [PubMed] [Google Scholar]
- 9.Blasig I, Löwe H, Ebert B (1987) Radical trapping and lipid peroxidation during myocardial reperfusion injury—radical scavenging by troxerutin in comparison to mannitol. Biomed Biochim Acta 46: S539–544. [PubMed] [Google Scholar]
- 10.Lu J, Wu D, Zheng Y, Hu B, Cheng W, Zhang Z, et al. (2013) Troxerutin counteracts domoic acid-induced memory deficits in mice by inhibiting CCAAT/enhancer binding protein β-mediated inflammatory response and oxidative stress. J Immunol 190: 3466–3479. doi: 10.4049/jimmunol.1202862 [DOI] [PubMed] [Google Scholar]
- 11.Kessler M, Ubeaud G, Walter T, Sturm F, Jung L (2002) Free radical scavenging and skin penetration of troxerutin and vitamin derivatives. J Dermatolog Treat 13: 133–141. doi: 10.1080/09546630260199505 [DOI] [PubMed] [Google Scholar]
- 12.Zhang Z, Fan S, Zheng Y, Lu J, Wu D, Shan Q, et al. (2009) Troxerutin protects the mouse liver against oxidative stress-mediated injury induced by D-galactose. J Agric Food Chem 57: 7731–7736. doi: 10.1021/jf9012357 [DOI] [PubMed] [Google Scholar]
- 13.Adam BS, Pentz R, Siegers CP, Strubelt O, Tegtmeier M (2005) Troxerutin protects the isolated perfused rat liver from a possible lipid peroxidation by coumarin. Phytomedicine 12: 52–61. doi: 10.1016/j.phymed.2004.01.007 [DOI] [PubMed] [Google Scholar]
- 14.Lu J, Wu DM, Hu B, Cheng W, Zheng YL, Zhang ZF, et al. (2010) Chronic administration of troxerutin protects mouse brain against D-galactose-induced impairment of cholinergic system. Neurobiol Learn Mem 93: 157–164. doi: 10.1016/j.nlm.2009.09.006 [DOI] [PubMed] [Google Scholar]
- 15.Chung H, Choi S, Ahn B, Kwak H, Kim J, Kim W (2005) Efficacy of troxerutin on streptozotocin-induced rat model in the early stage of diabetic retinopathy. Arzneimittelforschung 55: 573–580. doi: 10.1055/s-0031-1296907 [DOI] [PubMed] [Google Scholar]
- 16.Lu J, Wu DM, Zheng ZH, Zheng YL, Hu B, Zhang ZF (2011) Troxerutin protects against high cholesterol-induced cognitive deficits in mice. Brain 134: 783–797. doi: 10.1093/brain/awq376 [DOI] [PubMed] [Google Scholar]
- 17.Boisseau MR, Taccoen A, Garreau C, Vergnes C, Roudaut MF, Garreau-Gomez B (1995) Fibrinolysis and hemorheology in chronic venous insufficiency: a double blind study of troxerutin efficiency. J Cardiovasc Surg (Torino) 36: 369–374. [PubMed] [Google Scholar]
- 18.Gohel MS, Davies AH (2009) Pharmacological agents in the treatment of venous disease: an update of the available evidence. Curr Vasc Pharmacol 7: 303–308. [DOI] [PubMed] [Google Scholar]
- 19.Pyrzanowska J, Piechal A, Blecharz-Klin K, Joniec-Maciejak I, Zobel A, Widy-Tyszkiewicz E (2012) Influence of long-term administration of rutin on spatial memory as well as the concentration of brain neurotransmitters in aged rats. Pharmacol Rep 64: 808–816. [DOI] [PubMed] [Google Scholar]
- 20.Youdim KA, Dobbie MS, Kuhnle G, Proteggente AR, Abbott NJ, Rice-Evans C (2003) Interaction between flavonoids and the blood-brain barrier: in vitro studies. J Neurochem 85: 180–192. [DOI] [PubMed] [Google Scholar]
- 21.Conesa A, Madrigal P, Tarazona S, Gomez-Cabrero D, Cervera A, McPherson A, et al. (2016) A survey of best practices for RNA-seq data analysis. Genome Biol 17: 13 doi: 10.1186/s13059-016-0881-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang T, He K, Wang Z (2016) Transcriptome Comparison Analysis of Ostrinia furnacalis in Four Developmental Stages. Sci Rep 6: 35008 doi: 10.1038/srep35008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schefe JH, Lehmann KE, Buschmann IR, Unger T, Funke-Kaiser H (2006) Quantitative real-time RT-PCR data analysis: current concepts and the novel "gene expression's CT difference" formula. J Mol Med (Berl) 84: 901–910. [DOI] [PubMed] [Google Scholar]
- 24.Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111. doi: 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 doi: 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wagner GP, Kin K, Lynch VJ (2012) Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theory Biosci 131: 281–285. doi: 10.1007/s12064-012-0162-3 [DOI] [PubMed] [Google Scholar]
- 27.Liu W, Yang T, Yao C, Zuo S, Kong Y (2013) Spectroscopic Studies on the Interaction Between Troxerutin and Bovine Hemoglobin. Journal of Solution Chemistry 42: 1169–1182. [Google Scholar]
- 28.Maurya D, Salvi V, Nair K (2004) Radioprotection of Normal Tissues in Tumor-bearing Mice by Troxerutin. 221–228 p. [DOI] [PubMed] [Google Scholar]
- 29.Vinothkumar R, Vinoth Kumar R, Sudha M, Viswanathan P, Balasubramanian T, Nalini N (2014) Modulatory effect of troxerutin on biotransforming enzymes and preneoplasic lesions induced by 1,2-dimethylhydrazine in rat colon carcinogenesis. Exp Mol Pathol 96: 15–26. doi: 10.1016/j.yexmp.2013.10.009 [DOI] [PubMed] [Google Scholar]
- 30.Lee KS, Cha HJ, Lee GT, Lee KK, Hong JT, Ahn KJ, et al. (2014) Troxerutin induces protective effects against ultraviolet B radiation through the alteration of microRNA expression in human HaCaT keratinocyte cells. International Journal of Molecular Medicine 33: 934–942. doi: 10.3892/ijmm.2014.1641 [DOI] [PubMed] [Google Scholar]
- 31.Yu Y, Zheng G (2017) Troxerutin protects against diabetic cardiomyopathy through NF‑κB/AKT/IRS1 in a rat model of type 2 diabetes. Mol Med Rep 15: 3473–3478. doi: 10.3892/mmr.2017.6456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Geetha R, Radika M, Priyadarshini E, Bhavani K, Anuradha C (2015) Troxerutin reverses fibrotic changes in the myocardium of high-fat high-fructose diet-fed mice. Mol Cell Biochem 407: 263–279. doi: 10.1007/s11010-015-2474-3 [DOI] [PubMed] [Google Scholar]
- 33.Archimowicz-Cyry#owsk B, Adamek B, Droidzik M, Samochowiec L, Whjcicki J (1996) Clinical Effect of Buckwheat Herb, Ruscus Extract and Troxerutin on Retinopathy and Lipids in Diabetic Patients. PHYTOTHERAPY RESEARCH 10: 659–662. [Google Scholar]
- 34.Subastri A, Harikrishna K, Sureshkumar M, Alshammari G, Aristatile B, Thirunavukkarasu C (2017) Effect of troxerutin on 2-aminoanthracene and DNA interaction and its anti-mutagenic property. Biomed Pharmacother 88: 325–334. doi: 10.1016/j.biopha.2017.01.042 [DOI] [PubMed] [Google Scholar]
- 35.Jakovcevski I, Miljkovic D, Schachner M, Andjus P (2013) Tenascins and inflammation in disorders of the nervous system. Amino Acids 44: 1115–1127. doi: 10.1007/s00726-012-1446-0 [DOI] [PubMed] [Google Scholar]
- 36.El Ayachi I, Fernandez C, Baeza N, De Paula A, Pesheva P, Figarella-Branger D (2011) Spatiotemporal distribution of tenascin-R in the developing human cerebral cortex parallels neuronal migration. J Comp Neurol 519: 2379–2389. doi: 10.1002/cne.22632 [DOI] [PubMed] [Google Scholar]
- 37.Liao H, Bu WY, Wang TH, Ahmed S, Xiao ZC (2005) Tenascin-R plays a role in neuroprotection via its distinct domains that coordinate to modulate the microglia function. J Biol Chem 280: 8316–8323. doi: 10.1074/jbc.M412730200 [DOI] [PubMed] [Google Scholar]
- 38.Dufresne D, Hamdan FF, Rosenfeld JA, Torchia B, Rosenblatt B, Michaud JL, et al. (2012) Homozygous deletion of Tenascin-R in a patient with intellectual disability. J Med Genet 49: 451–454. doi: 10.1136/jmedgenet-2012-100831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saghatelyan A, de Chevigny A, Schachner M, Lledo PM (2004) Tenascin-R mediates activity-dependent recruitment of neuroblasts in the adult mouse forebrain. Nat Neurosci 7: 347–356. doi: 10.1038/nn1211 [DOI] [PubMed] [Google Scholar]
- 40.Freitag S, Schachner M, Morellini F (2003) Behavioral alterations in mice deficient for the extracellular matrix glycoprotein tenascin-R. Behav Brain Res 145: 189–207. [DOI] [PubMed] [Google Scholar]
- 41.Montag-Sallaz M, Montag D (2003) Severe cognitive and motor coordination deficits in tenascin-R-deficient mice. Genes Brain Behav 2: 20–31. [DOI] [PubMed] [Google Scholar]
- 42.Gurevicius K, Gureviciene I, Valjakka A, Schachner M, Tanila H (2004) Enhanced cortical and hippocampal neuronal excitability in mice deficient in the extracellular matrix glycoprotein tenascin-R. Mol Cell Neurosci 25: 515–523. doi: 10.1016/j.mcn.2003.12.001 [DOI] [PubMed] [Google Scholar]
- 43.Anlar B, Gunel-Ozcan A (2012) Tenascin-R: role in the central nervous system. Int J Biochem Cell Biol 44: 1385–1389. doi: 10.1016/j.biocel.2012.05.009 [DOI] [PubMed] [Google Scholar]
- 44.Lu J, Wu D, Hu B, Cheng W, Zheng Y, Zhang Z, et al. (2010) Chronic administration of troxerutin protects mouse brain against D-galactose-induced impairment of cholinergic system. Neurobiol Learn Mem 93: 157–164. doi: 10.1016/j.nlm.2009.09.006 [DOI] [PubMed] [Google Scholar]
- 45.Zhang S, Li H, Zhang L, Li J, Wang R, Wang M (2017) Effects of troxerutin on cognitive deficits and glutamate cysteine ligase subunits in the hippocampus of streptozotocin-induced type 1 diabetes mellitus rats. Brain Res 1657: 355–360. doi: 10.1016/j.brainres.2016.12.009 [DOI] [PubMed] [Google Scholar]
- 46.Tatemoto K, Carlquist M, Mutt V (1982) Neuropeptide Y—a novel brain peptide with structural similarities to peptide YY and pancreatic polypeptide. Nature 296: 659–660. [DOI] [PubMed] [Google Scholar]
- 47.Zhou Z, Zhu G, Hariri A, Enoch M, Scott D, Sinha R, et al. (2008) Genetic variation in human NPY expression affects stress response and emotion. Nature 452: 997–1001. doi: 10.1038/nature06858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu Y, Andiappan A, Lee B, Ho R, Lim T, Kuan W, et al. (2016) Neuropeptide Y associated with asthma in young adults. Neuropeptides 59: 117–121. doi: 10.1016/j.npep.2016.07.003 [DOI] [PubMed] [Google Scholar]
- 49.Michel M, Beck-Sickinger A, Cox H, Doods H, Herzog H, Larhammar D, et al. (1998) XVI. International Union of Pharmacology recommendations for the nomenclature of neuropeptide Y, peptide YY, and pancreatic polypeptide receptors. Pharmacol Rev 50: 143–150. [PubMed] [Google Scholar]
- 50.Wheway J, Mackay C, Newton R, Sainsbury A, Boey D, Herzog H, et al. (2005) A fundamental bimodal role for neuropeptide Y1 receptor in the immune system. J Exp Med 202: 1527–1538. doi: 10.1084/jem.20051971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schwarz H, Villiger P, von Kempis J, Lotz M (1994) Neuropeptide Y is an inducible gene in the human immune system. J Neuroimmunol 51: 53–61. [DOI] [PubMed] [Google Scholar]
- 52.Macia L, Yulyaningsih E, Pangon L, Nguyen A, Lin S, Shi Y, et al. (2012) Neuropeptide Y1 receptor in immune cells regulates inflammation and insulin resistance associated with diet-induced obesity. Diabetes 61: 3228–3238. doi: 10.2337/db12-0156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Farzi A, Reichmann F, Holzer P (2015) The homeostatic role of neuropeptide Y in immune function and its impact on mood and behaviour. Acta Physiol (Oxf) 213: 603–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pang X, Li T, Xie Q, He F, Cui D, Chen Y, et al. (2010) Amelioration of dextran sulfate sodium-induced colitis by neuropeptide Y antisense oligodeoxynucleotide. Int J Colorectal Dis 25: 1047–1053. doi: 10.1007/s00384-010-0964-z [DOI] [PubMed] [Google Scholar]
- 55.Holzer P, Reichmann F, Farzi A (2012) Neuropeptide Y, peptide YY and pancreatic polypeptide in the gut-brain axis. Neuropeptides 46: 261–274. doi: 10.1016/j.npep.2012.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Geetha R, Yogalakshmi B, Sreeja S, Bhavani K, Anuradha C (2014) Troxerutin suppresses lipid abnormalities in the heart of high-fat-high-fructose diet-fed mice. Mol Cell Biochem 387: 123–134. doi: 10.1007/s11010-013-1877-2 [DOI] [PubMed] [Google Scholar]
- 57.Pelava A, Schneider C, Watkins N (2016) The importance of ribosome production, and the 5S RNP-MDM2 pathway, in health and disease. Biochem Soc Trans 44: 1086–1090. doi: 10.1042/BST20160106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gamalinda M, Woolford J (2015) Paradigms of ribosome synthesis: Lessons learned from ribosomal proteins. Translation (Austin) 3: e975018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang W, Nag S, Zhang X, Wang M, Wang H, Zhou J, et al. (2015) Ribosomal proteins and human diseases: pathogenesis, molecular mechanisms, and therapeutic implications. Med Res Rev 35: 225–285. doi: 10.1002/med.21327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gamalinda M, Jakovljevic J, Babiano R, Talkish J, de la Cruz J, Woolford J (2013) Yeast polypeptide exit tunnel ribosomal proteins L17, L35 and L37 are necessary to recruit late-assembling factors required for 27SB pre-rRNA processing. Nucleic Acids Res 41: 1965–1983. doi: 10.1093/nar/gks1272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smolock E, Korshunov V, Glazko G, Qiu X, Gerloff J, Berk B (2012) Ribosomal protein L17, RpL17, is an inhibitor of vascular smooth muscle growth and carotid intima formation. Circulation 126: 2418–2427. doi: 10.1161/CIRCULATIONAHA.112.125971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Meskauskas A, Dinman J (2007) Ribosomal protein L3: gatekeeper to the A site. Mol Cell 25: 877–888. doi: 10.1016/j.molcel.2007.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kobayashi Y, Kawakami K, Ohbayashi M, Kohyama N, Yamamoto T (2010) Ribosomal protein L3 mediated the transport of digoxin in Xenopus laevis oocyte. J Toxicol Sci 35: 827–834. [DOI] [PubMed] [Google Scholar]
- 64.Russo A, Saide A, Cagliani R, Cantile M, Botti G, Russo G (2016) rpL3 promotes the apoptosis of p53 mutated lung cancer cells by down-regulating CBS and NFκB upon 5-FU treatment. Sci Rep 6: 38369 doi: 10.1038/srep38369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ohkia A, Hu Y, Wang M, Garcia F, Stearns M (2004) Evidence for prostate cancer-associated diagnostic marker-1: immunohistochemistry and in situ hybridization studies. Clin Cancer Res 10: 2452–2458. [DOI] [PubMed] [Google Scholar]
- 66.Lütcke H (1995) Signal recognition particle (SRP), a ubiquitous initiator of protein translocation. Eur J Biochem 228: 531–550. [DOI] [PubMed] [Google Scholar]
- 67.Bacher G, Lütcke H, Jungnickel B, Rapoport T, Dobberstein B (1996) Regulation by the ribosome of the GTPase of the signal-recognition particle during protein targeting. Nature 381: 248–251. doi: 10.1038/381248a0 [DOI] [PubMed] [Google Scholar]
- 68.Birse D, Kapp U, Strub K, Cusack S, Aberg A (1997) The crystal structure of the signal recognition particle Alu RNA binding heterodimer, SRP9/14. EMBO J 16: 3757–3766. doi: 10.1093/emboj/16.13.3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rock K, Reits E, Neefjes J (2016) Present Yourself! By MHC Class I and MHC Class II Molecules. Trends Immunol 37: 724–737. doi: 10.1016/j.it.2016.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Flajnik M, Kasahara M (2010) Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat Rev Genet 11: 47–59. doi: 10.1038/nrg2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Garcia-Lora A, Algarra I, Garrido F (2003) MHC class I antigens, immune surveillance, and tumor immune escape. J Cell Physiol 195: 346–355. doi: 10.1002/jcp.10290 [DOI] [PubMed] [Google Scholar]
- 72.Smit E, Baas P (2016) Lung cancer in 2015: Bypassing checkpoints, overcoming resistance, and honing in on new targets. Nat Rev Clin Oncol 13: 75–76. doi: 10.1038/nrclinonc.2015.223 [DOI] [PubMed] [Google Scholar]
- 73.Endo Y, Nakazawa N, Liu Y, Iwaki D, Takahashi M, Fujita T, et al. (2005) Carbohydrate-binding specificities of mouse ficolin A, a splicing variant of ficolin A and ficolin B and their complex formation with MASP-2 and sMAP. Immunogenetics 57: 837–844. doi: 10.1007/s00251-005-0058-1 [DOI] [PubMed] [Google Scholar]
- 74.Endo Y, Matsushita M, Fujita T (2007) Role of ficolin in innate immunity and its molecular basis. Immunobiology 212: 371–379. doi: 10.1016/j.imbio.2006.11.014 [DOI] [PubMed] [Google Scholar]
- 75.Matsushita M, Fujita T (2002) The role of ficolins in innate immunity. Immunobiology 205: 490–497. doi: 10.1078/0171-2985-00149 [DOI] [PubMed] [Google Scholar]
- 76.Holt CB, Ostergaard JA, Axelgaard E, Nielsen GK, Endo Y, Thiel S, et al. (2015) Ficolin B in Diabetic Kidney Disease in a Mouse Model of Type 1 Diabetes. Mediators Inflamm 2015: 653260 doi: 10.1155/2015/653260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ren Y, Ding Q, Zhang X (2014) Ficolins and infectious diseases. Virol Sin 29: 25–32. doi: 10.1007/s12250-014-3421-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ypsilanti A, Zagar Y, Chédotal A (2010) Moving away from the midline: new developments for Slit and Robo. Development 137: 1939–1952. doi: 10.1242/dev.044511 [DOI] [PubMed] [Google Scholar]
- 79.Macias H, Moran A, Samara Y, Moreno M, Compton J, Harburg G, et al. (2011) SLIT/ROBO1 signaling suppresses mammary branching morphogenesis by limiting basal cell number. Dev Cell 20: 827–840. doi: 10.1016/j.devcel.2011.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chang J, Lan T, Li C, Ji X, Zheng L, Gou H, et al. (2015) Activation of Slit2-Robo1 signaling promotes liver fibrosis. J Hepatol 63: 1413–1420. doi: 10.1016/j.jhep.2015.07.033 [DOI] [PubMed] [Google Scholar]
- 81.Huang L, Xu Y, Yu W, Li Y, Chu L, Dong J, et al. (2010) Effect of Robo1 on retinal pigment epithelial cells and experimental proliferative vitreoretinopathy. Invest Ophthalmol Vis Sci 51: 3193–3204. doi: 10.1167/iovs.09-3779 [DOI] [PubMed] [Google Scholar]
- 82.Li J, Ye Y, Zhang R, Zhang L, Hu X, Han D, et al. (2015) Robo1/2 regulate follicle atresia through manipulating granulosa cell apoptosis in mice. Sci Rep 5: 9720 doi: 10.1038/srep09720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rajagopalan G, Chandrasekaran S, Carani Venkatraman A (2017) Troxerutin attenuates diet-induced oxidative stress, impairment of mitochondrial biogenesis and respiratory chain complexes in mice heart. Clin Exp Pharmacol Physiol 44: 103–113. doi: 10.1111/1440-1681.12671 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
(XLS)
(XLS)
(XLS)
(XLS)
(XLS)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.