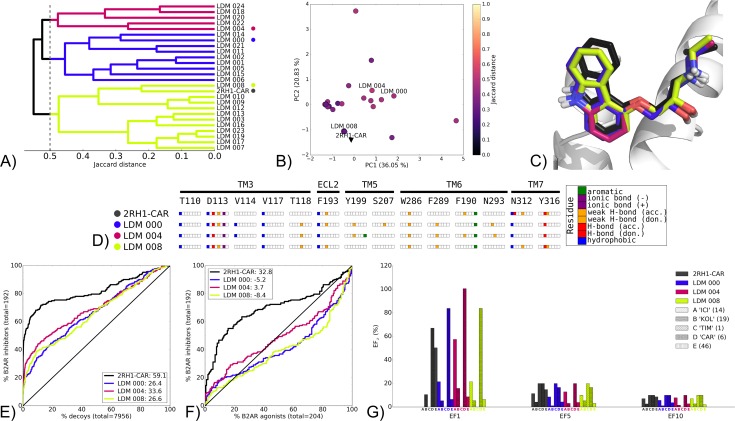
Fig 4. Self refinement LDM on the B2AR 2RH1-CAR, using CAR as the refinement ligand.
A) Dendrogram of the top 25 LDM models and the X-ray structure, a cutoff line identifies different LDM clusters. Representative LDM models are the highest scoring within the cluster based on the OPUS-ICM metric and are designated by a coloured dot. B) Comparison of binding pocket conformation between the top 25 LDM models and the X-ray structure. LDM models are colored based on their IFP Jaccard distance with the destination X-ray structure, 2RH1-CAR. C) Binding poses of the representative LDM models and the destination X-ray structure with the X-ray ligand in black and the LDM models coloured based on their representative clusters defined in A. D) IFP of the representative LDM models and the X-ray structure. Interaction type is described for each residue of the binding pocket: hydrophobic interaction, hydrogen bond (H-bond) donor and acceptor, weak hydrogen bond (weak H-bond) donor and acceptor, ionic bond positive (+) and negative (-) and aromatic interaction. VS performance is described with ROC curves to visualise E) the recovery of B2AR inhibitors vs. decoys and F) the selectivity of B2AR inhibitors over B2AR agonists. X-ray structures are in black, with the LDM models coloured based on their relative clusters identified in A. The relative rank of the LDM refinement ligand is identified with a vertical dashed line. In all cases, this vertical line is masked by the axis indicating that the refinement ligand was very highly ranked. The ROC curve figure inset shows NSQ_AUC values for each binding pocket. G) bar chart is used to visualise the EF for representative B2AR inhibitor chemotypes at EF1, EF5 and EF10. Chemotypes A ‘ICI-like’, B ‘KOL-like’, C ‘TIM-like’, D ‘CAR-like’ and E represent only a subset of B2AR inhibitor ligands (S2D Fig). The EF bar chart inset shows the number of ligands for each chemotype cluster between parenthesis. X-ray structure chemotype EF shown in black bars, with the LDM models coloured based on their relative clusters identified in A.