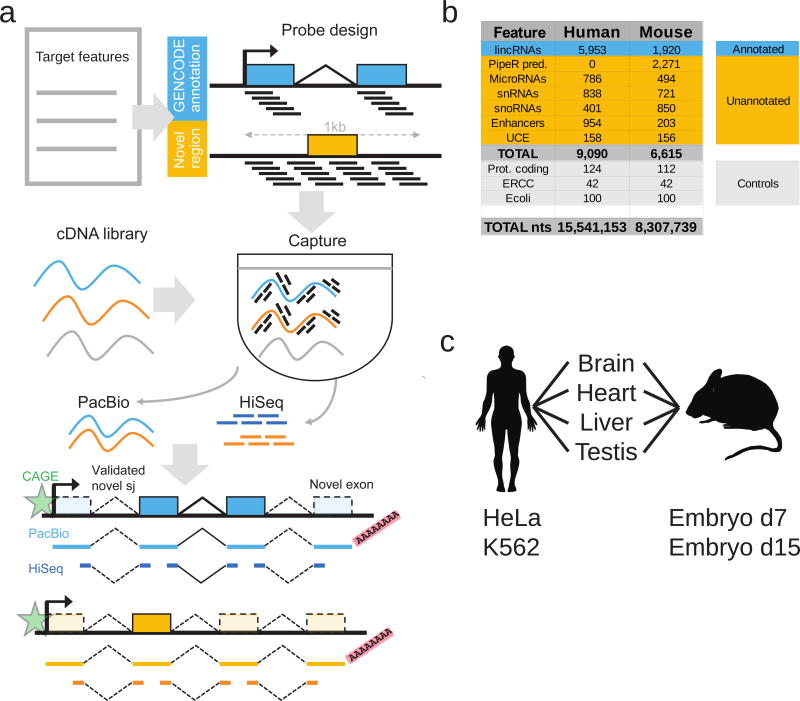
Figure 1. Capture Long Seq approach to extend the GENCODE lncRNA annotation.
a) Strategy for automated, high-quality transcriptome annotation. CLS may be used to complete existing annotations (blue), or to map novel transcript structures in suspected loci (orange). Capture oligonucleotides (black bars) are designed to tile across targeted regions. PacBio libraries are prepared for from the captured molecules. Illumina HiSeq short-read sequencing can be performed for independent validation of predicted splice junctions. Predicted transcription start sites can be confirmed by CAGE clusters (green), and transcription termination sites by non-genomically encoded polyA sequences in PacBio reads. Novel exons are denoted by lighter coloured rectangles.
b) Summary of human and mouse capture library designs. Shown are the number of individual gene loci that were probed. “PipeR pred.”: orthologue predictions in mouse genome of human lncRNAs, made by PipeR29; “UCE”: ultraconserved elements; “Prot. coding”: expression-matched, randomly-selected protein-coding genes; “ERCC”: spike-in sequences; “Ecoli”: randomly-selected E. coli genomic regions. Enhancers and UCEs are probed on both strands, and these are counted separately. “Total nts”: sum of targeted nucleotides.
c) RNA samples used.