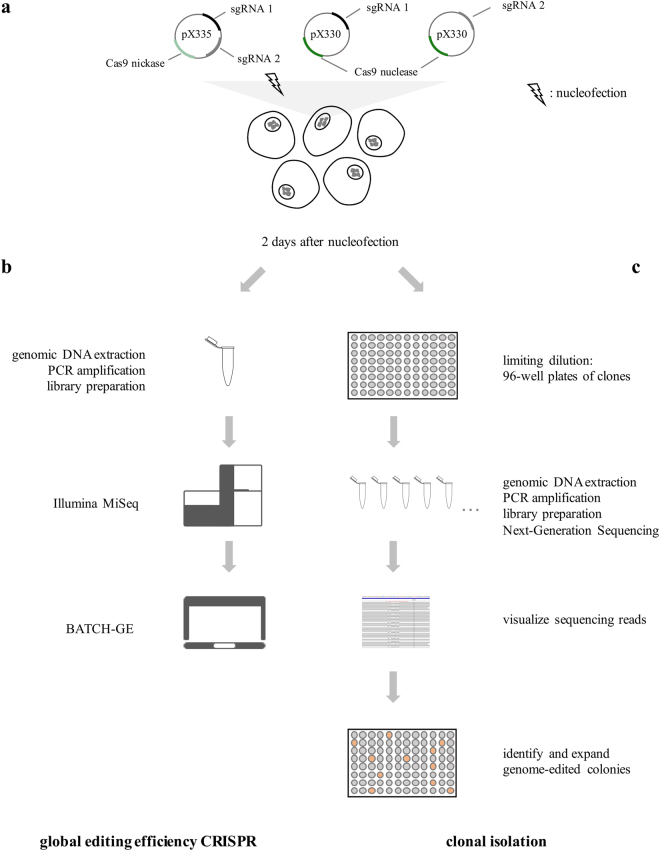
Figure 1.
Optimized workflow for efficient genome-editing of naïve human embryonic stem cells. (a) Two sgRNAs were designed for each gene to allow double nicking and cloned in the pX335 plasmid. Both sgRNAs were also individually paired with the Cas9 nuclease (pX330 plasmid). The CRISPR plasmids were transfected in the naïve hESCs using nucleofection at day two after splitting. (b) To determine the overall editing efficiency of the CRISPR, DNA was extracted from the pool of transfected cells two days after nucleofection. After subsequent PCR and massive parallel sequencing (Illumina MiSeq), the overall editing efficiency and the detected indel variants were quantified with BATCH-GE51. (c) To obtain monoclonal genome-edited colonies, clonal isolation was performed two days after nucleofection, using limiting dilution (0.5 cells/100 µl). After 6–8 days, surviving clones were observed and DNA was extracted. After amplification and massive parallel sequencing of the target region, the sequencing reads were visualized in the Integrative Genomics Viewer (IGV, Broad Institute) to identify the genome-edited colonies.