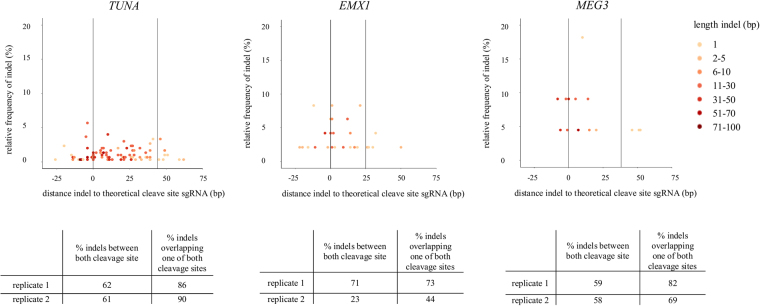
Figure 4.
Majority of indels is present between both cleavage sites after editing with Cas9 nickase. Distance of the start position of each insertion or deletion to the theoretical cleavage site (=3 bp upstream of the PAM sequence) is presented on the x-axis, while the relative frequency of the indel is presented on the y-axis after editing with the Cas9 nickase. Length of indels are represented by the different colors. Only 1 replicate is shown. The majority of indels is present between the two theoretical cleavage sites (indicated by the black lines at position 0 (sgRNA1) and at positions 44 bp (TUNA), 25 bp (EMX1) and 38 bp (MEG3). Percentages of indels 1) within or spanning both the two cleavage sites and 2) indels spanning at least one of the two sites are shown below the graph. Replicate 2 of EMX1 has an editing efficiency of 2.7% (compared to 5% for replicate 1), which can explain the lower percentage of indels located between the two cleavage sites.