Figure 2.
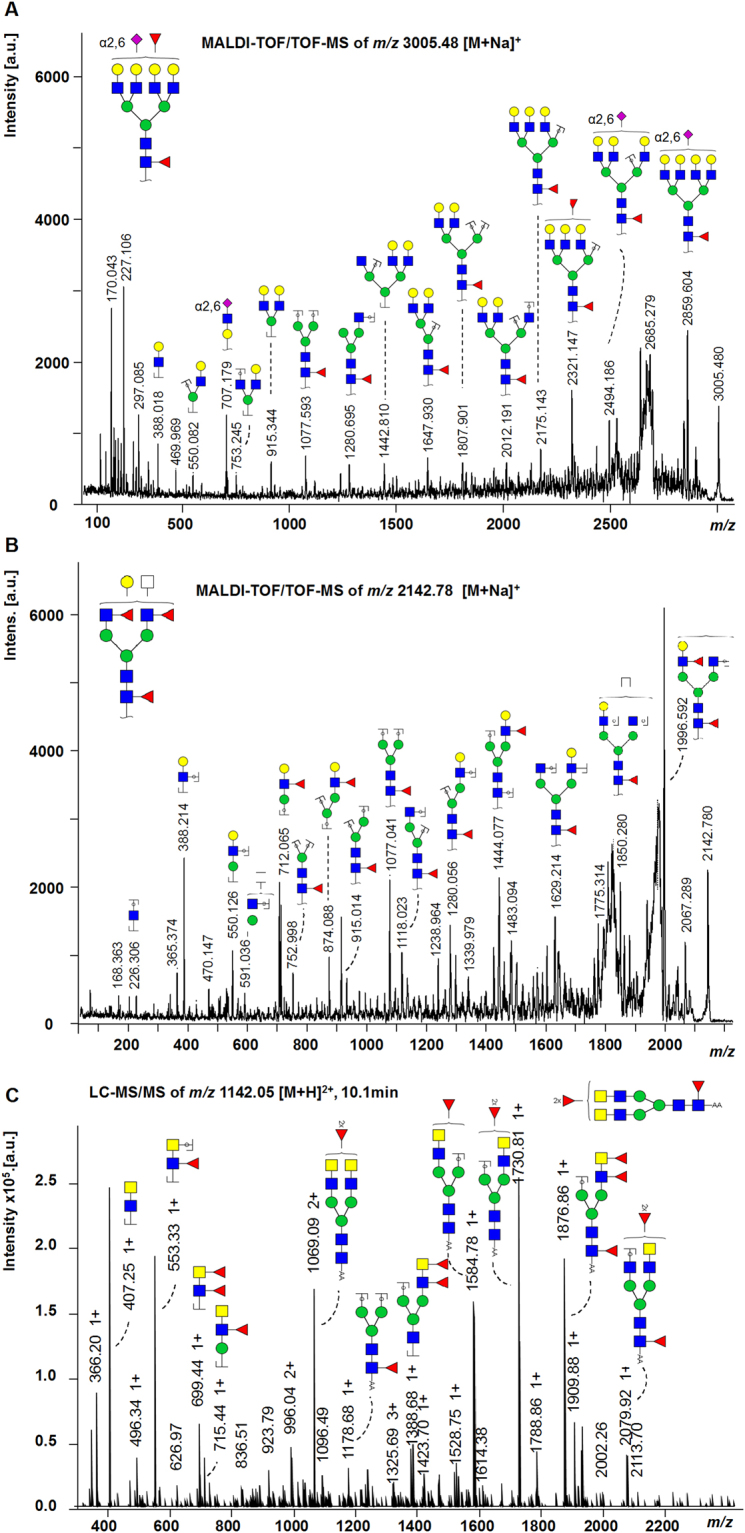
Fragmentation spectra. (A) MALDI-TOF/TOF-MS/MS fragmentation spectrum of the N-glycan Hex7HexNAc6Fuc2(α2,6)NeuAc1 with m/z 3005.48 [M + Na]+. The fragment ion at m/z 707.2 [M + Na]+ is indicative for Hex1HexNAc1(α2,6)NeuAc1. The mass shift of + 28 Da from a non-modified N-acetylneuraminic acid to an ethyl esterified N-acetylneuraminic acid indicates α2,6-linkage. The position of the α2,6NeuAc as well as the antenna fucose cannot be identified. (B) MALDI-TOF/TOF-MS/MS fragmentation spectrum of the N-glycan Hex4HexNAc5Fuc3 with m/z 2142.78 [M + Na]+. Fragment ions for antenna-fucosylation (m/z 712.1 [M + Na]+, m/z 874.1 [M + Na]+) as well as core-fucosylation (m/z 1077.0 [M + Na]+) were identified. (C) LC-MS/MS fragmentation spectrum of the N-glycan Hex3 HexNAc6Fuc3 with m/z 1142.05 [M + H]2+. Indicative fragment ions at m/z 407 [M + H]+ (HexNAc2) and m/z 553 [M + H]+ (HexNAc2dHex1) show the presence of LacdiNAc structures. Annotation was performed in GlycoWorkbench 2.1 stable build 146 (http://www.eurocarbdb.org/) using the Glyco-Peakfinder tool (http://www.eurocarbdb.org/ms-tools/). The presence of structural isomers cannot be excluded. Hex = hexose; blue circle = Glc, glucose; yellow circle = Gal, galactose; green circle = Man, mannose; blue square = GlcNAc, N-acetylglucosamine; yellow square = GalNAc, N-acetylgalactosamine; white square = HexNAc, N-acetylhexosamine; red triangle = Fuc, deoxyhexose, fucose; purple diamond = NeuAc, N-acetylneuraminic acid; *reducing end adduct.
