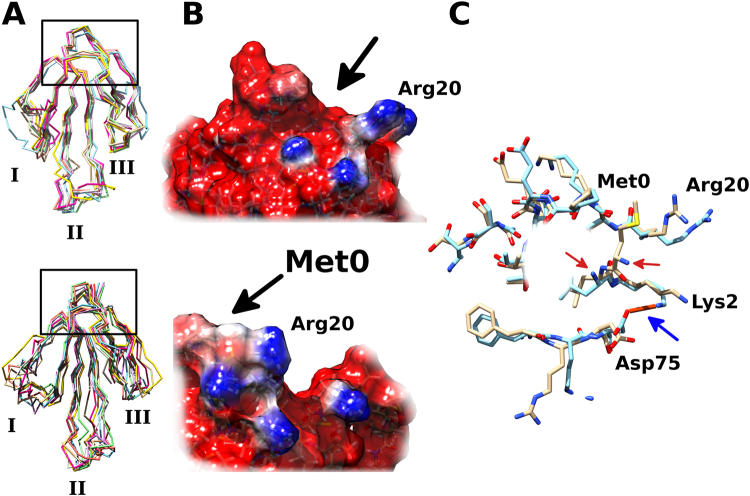
Figure 7.
Comparison of the synthetic and recombinant human SLURP-1 structural models. (A) Deletion of the methionine (Met0) residue did not significantly alter the overall molecule motility of sSLURP-1 (top). For comparison, rSLURP-1 (bottom) is also presented. Seven superimposed frames from molecular dynamics simulations are shown. The N-terminal region of interest is boxed and the three protruding fingers are labeled I–III. (B) Electrostatic-surface profile of rSLURP-1 (bottom) and sSLURP-1 (top) showing that the positively charged Arg20 is now more solvent exposed. Arrows show regions occupied by Met0 residue in rSLURP-1 or solvent-accessible area in sSLURP-1. (C) Superimposed rSLURP-1 (light brown) and sSLURP-1 (cyan) structures showing the differences in the “head” region where Met0 is located in rSLURP-1. The N-terminal amino groups are indicated by red arrows. In the sSLURP-1 structure, a salt-bridge between residues Lys2 and Asp75 is present (blue arrow). Some residues have been omitted for clarity.