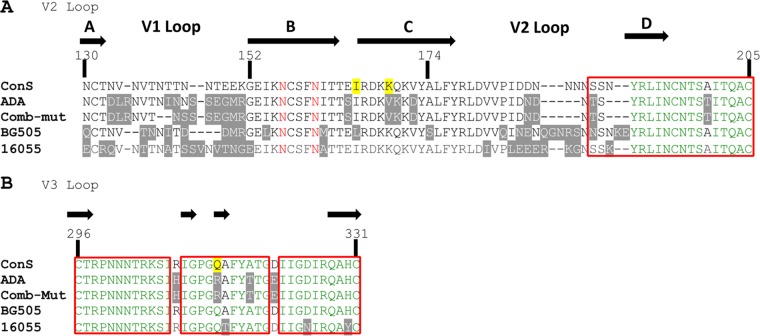
FIG 2.
Alignment of V1V2 and V3 sequences. The V1V2 (A) and V3 (B) amino acid sequences of the group M consensus (ConS), ADA, Comb-mut, BG505, and 16055 were aligned using DNASTAR Lasergene ClustalW with an HxB2 numbering scheme. Highlighted in gray are residues that differ from ConS. Residues in red indicate N-linked glycosylation sites at N156 and N160. Highlighted in yellow are positions 165, 169, and 315; the former two were mutated I165L/V169K to generate ADA-LK and CM-LK, and the latter was mutated R315Q to eliminate neutralization by V3 crown antibodies. Amino acids represented in green are >50% conserved among all isolates, and those boxed in red are >50% conserved among subtype B (hiv.lanl.org). The beta-sheet structure and labeling are indicated by black arrows, based on data published by McLellan et al. for V2 (21) and Huang et al. for V3 (87).