FIG 4.
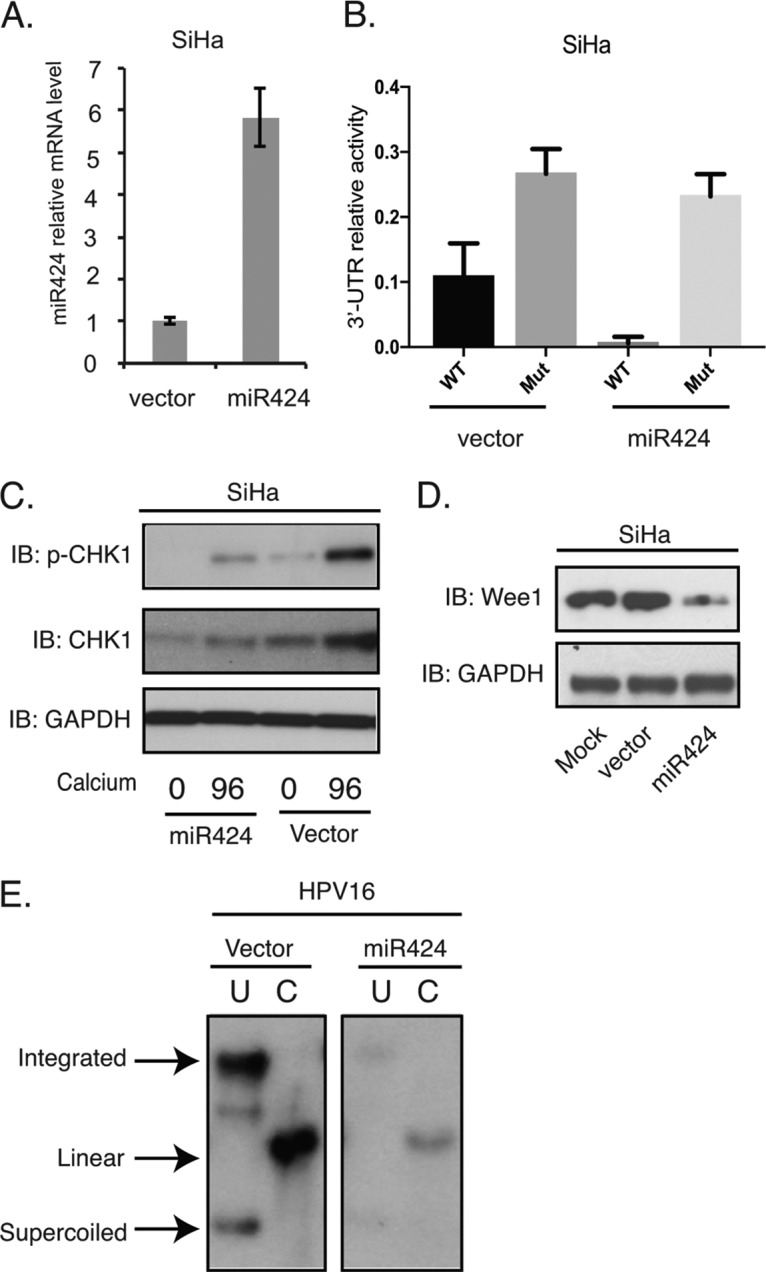
The suppression of miR-424 is required for HPV16 genome maintenance. (A) RT-PCR assay of miR-424 mRNA levels in SiHa cells transduced with lentiviruses expressing the control vector or miR-424 (P < 0.03). (B) The luciferase reporter vector containing wild-type CHK1 3′ UTR or a mutant CHK1 3′ UTR were transfected into SiHa cells transduced with miR424-expressing lentiviruses. The firefly luciferase activities were determined 48 h after transfection and normalized to Renilla luciferase. The relative luciferase activities were normalized to the control pmiRGLO plasmid. Each column stands for the mean value plus the standard deviation of three independent experiments. (C) Western blot analysis of p-CHK1, CHK1, and GAPDH proteins in SiHa cells transduced with the control and miR-424 lentiviruses, followed by differentiation in high-calcium media for 96 h. (D) Western blot analysis of Wee1 and GAPDH proteins in undifferentiated SiHa cells transduced with the control and miR-424 lentiviruses. (E) Southern blot analysis for HPV16 episomes in the control or miR-424-expressing HPV16 cells. The total DNAs from cells grown in monolayers were incubated with an enzyme that does not digest the HPV16 genome (lanes U) or cuts it once (lanes C). All results are representative of observations from three independent experiments.
