Figure 4. Molecular requirements for COMA assembly.
-
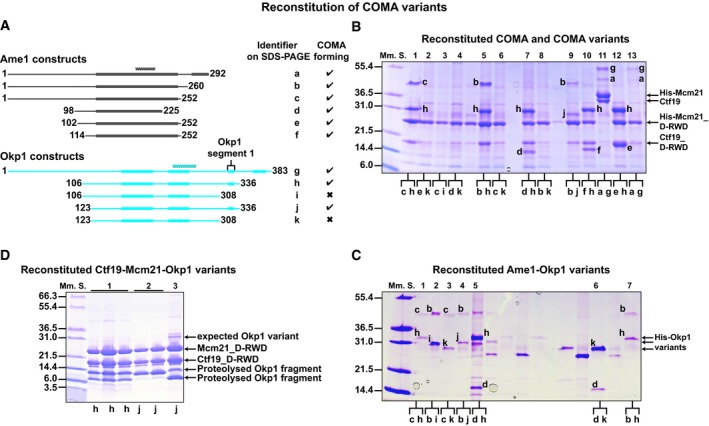
ASchematics of our designed Ame1 and Okp1 constructs; our identified structured segments and coiled coils are as we show in Fig 3.
-
B, CRepresentative images of Coomassie Blue‐stained SDS–PAGE gels with Ni2+‐affinity purification eluates of (B) full‐length COMA (lane 11) with full‐length polyhistidine‐tagged Mcm21 (His‐Mcm21) and Ctf19, or COMA variants with polyhistidine‐tagged Mcm21 D‐RWD domain (His‐Mcm21_D‐RWD; K. lactis Mcm21 residues 108–293) and Ctf19 D‐RWD domain (Ctf19_D‐RWD; K. lactis Ctf19 residues 107–270); of (C) Ame1‐Okp1 variants with polyhistidine‐tagged Okp1 variants (His‐Okp1 variants). We show one‐letter identifiers for protein versions in (A). Lanes between lanes 5 and 6 in (C) are not relevant.
-
DImage of SDS–PAGE gel with fractions of two purified Ctf19‐Mcm21‐Okp1 variants (after SEC); expected migrating position of non‐proteolysed Okp1 variant (compare with its position in SDS–PAGE images of COMA or Ame1‐Okp1 variants in (B or C)) is marked; 2 and 3: SEC‐purified fractions of samples from different elution fractions from ion‐exchange chromatography. When we co‐expressed full‐length Okp1 with Ctf19 and Mcm21, which has a coding region for an N‐terminal polyhistidine tag, we did not co‐purify substantial amounts of full‐length Okp1 with Ctf19‐Mcm21, probably because Okp1 was prone to proteolysis (in the absence of Ame1).
