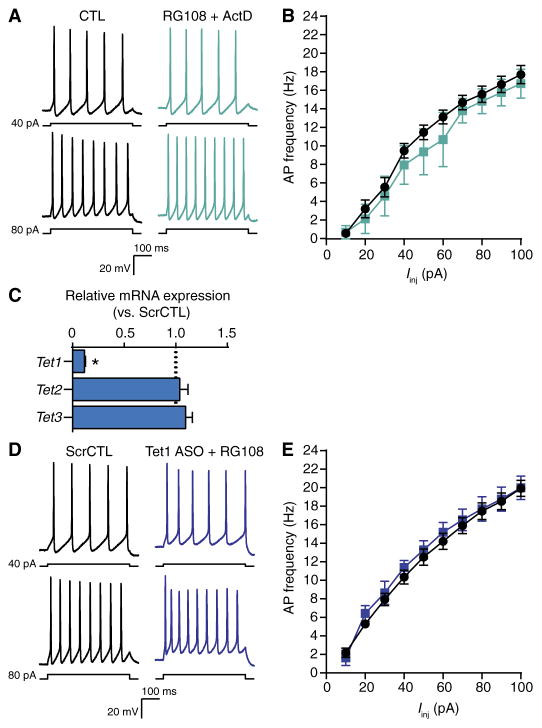
Fig. 3. Gene expression and DNA demethylation mediated by TET1 are necessary for DNMTi-enhanced excitability.
(A) Representative recordings of voltage responses from pyramidal neurons after 24-hour exposure to CTL or RG108 + actinomycin D. (B) Mean AP firing rates evoked from increasing current injections. Graph shows means ± SEM from cells pooled from at least three experiments for each condition (CTL, n = 11 cells; RG108 + actinomycin D, n = 4 cells). (C) Relative expression versus scrambled control of Tet1, Tet2, and Tet3 mRNA after treatment with a Tet1-targeting ASO. Data are normalized to the abundance in the scrambled control and are means ± SEM from three experiments (ScrCTL, n = 12 biological replicates; Tet1 ASO, n = 9 biological replicates). *P < 0.05, Mann-Whitney test. (D) Representative evoked membrane voltage responses of cells exposed to scrambled control ASO or Tet1 ASO + 24-hour RG108. (E) Mean AP firing rates evoked with 500-ms current pulses of increasing intensity. Bar graph shows means ± SEM from cells pooled from at least three experiments for each condition (scrambled control, n = 6 cells; Tet1 ASO + RG108, n = 7 cells).