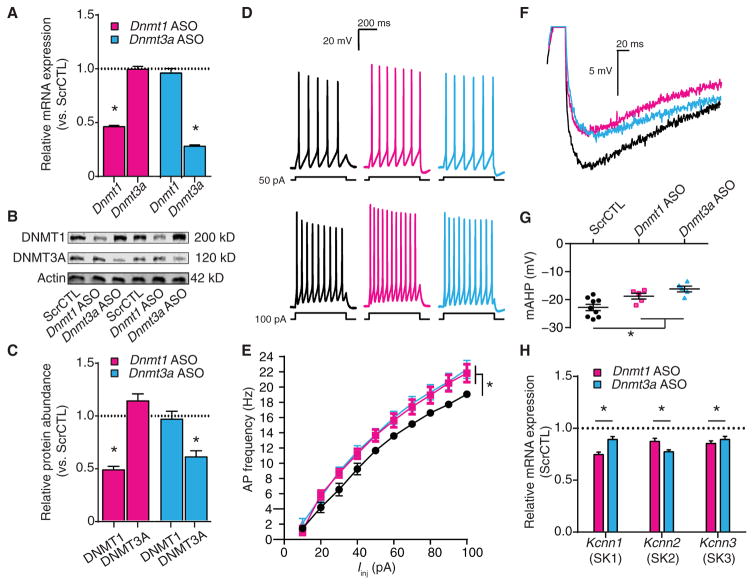
Fig. 6. DNMT1 or DNMT3A knockdown increases excitability, decreases the mAHP, and decreases expression of SK channel–encoding genes.
Neurons were exposed to scrambled control, Dnmt1 ASO, or Dnmt3a ASO. (A) Relative Dnmt1 and Dnmt3a mRNA expression after ASO exposure (scrambled control, n = 9 biological replicates; Dnmt1 ASO, n = 9 biological replicates; Dnmt3a ASO, n = 9 biological replicates). (B and C) Representative immunoblot (B) and relative DNMT1 and DNMT3A protein abundance (C) after ASO exposure (scrambled control, n = 9 biological replicates; Dnmt1 ASO, n = 9 biological replicates; Dnmt3a ASO, n = 8 biological replicates). (D and E) Representative recordings of evoked firing responses (D) and mean firing rates (E) of ASO-treated neurons (scrambled control, n = 12 cells; Dnmt1 ASO, n = 9 cells; Dnmt3a ASO, n = 7 cells). (F) Representative recordings of single APs from ASO-treated pyramidal neurons overlaid to show the mAHP. (G) Graph of mAHP amplitudes after ASO exposure (scrambled control, n = 9 cells; Dnmt1 ASO, n = 5 cells; Dnmt3a ASO, n = 5 cells). (H) Relative transcript expression of the SK channel genes Kcnn1, Kcnn2, and Kcnn3 after ASO exposure (scrambled control, n = 9 biological replicates; Dnmt1 ASO, n = 9 biological replicates; Dnmt3a ASO, n = 9 biological replicates). (A, C, and H) Data are normalized to scrambled control and are means ± SEM from three experiments. (E and G) Graphs show means ± SEM from cells pooled from at least three experiments for each condition. (A, C, G, and H) *P < 0.05, one-way ANOVA followed by Dunnett’s test. (E) *P < 0.05, RM-ANOVA.