Figure 4.
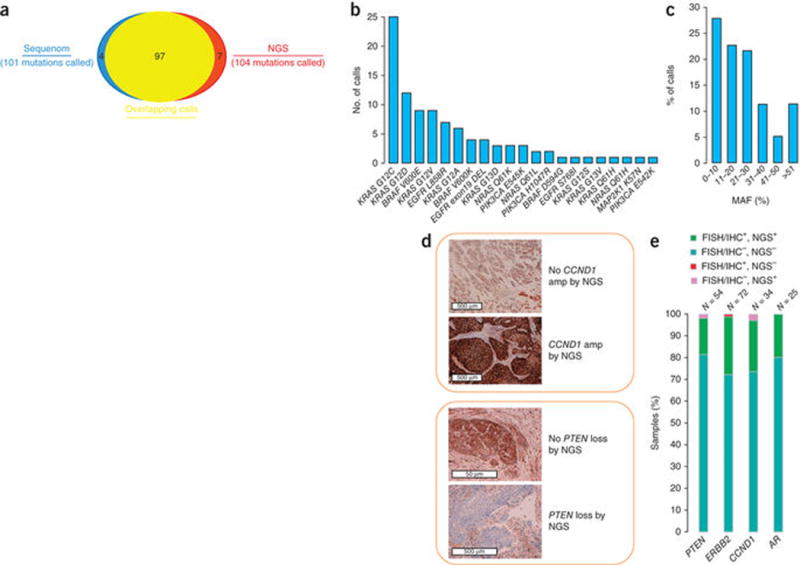
Concordance with clinical testing on FFPE specimens. Tumor specimens (N = 249) were assayed using the NGS-based test and by several other methods. (a) Overlap between positive alteration calls by NGS and Sequenom or gel sizing at 91 mutually tested sites in 118 FFPE clinical cancer specimens. (b) Specific alterations comprising the 97 concordant calls. (c) Histogram of NGS MAF in the 97 concordant calls. The high prevalence of low MAF in clinical cancer specimens is highlighted. (d) Examples of confirmation of NGS CNAs by IHC. (e) Summary of concordance between NGS, FISH and IHC findings across four genes (Supplementary Table 12).
