FIGURE 1.
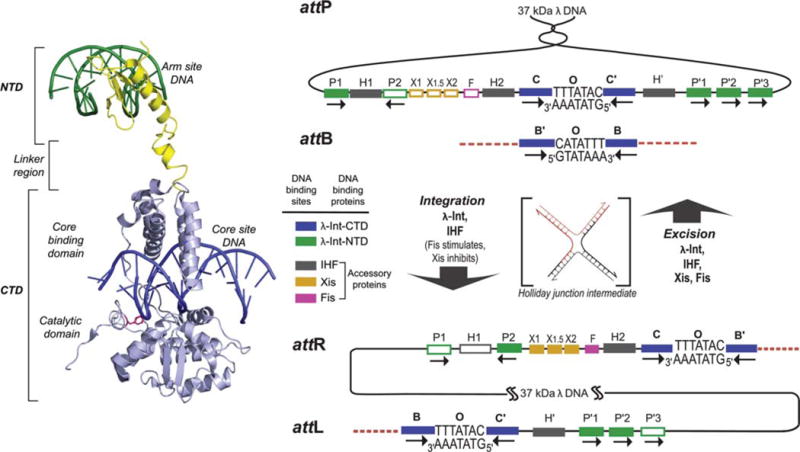
λ Integrase and the overlapping ensembles of protein binding sites that comprise att site DNA. The left panel shows the structure of a single λ Int protomer bound via its NTD to an arm site DNA and via its CTD to a core site DNA (adapted from the Int tetrameric structure determined by Biswas et al. [44], PDB code 1Z1G). The right panel shows the recombination reactions. Integrative recombination between supercoiled attP and linear attB requires the virally encoded integrase (Int) (2) and the host-encoded accessory DNA bending protein integration host factor (IHF) (4,177) and gives rise to an integrated phage chromosome bounded by attL and attR. Excisive recombination between attL and attR to regenerate attP and attB additionally requires the phage-encoded Xis protein (which inhibits integrative recombination) (140) and is stimulated by the host-encoded Fis protein (8). Both reactions proceed through a Holliday junction intermediate that is first generated and then resolved by single strand exchanges on the left and right side of the 7 bp overlap region, respectively. The two reactions proceed with the same order of sequential strand exchanges (not the reverse order) and use different subsets of protein binding sites in the P and P′ arms, as indicated by the filled boxes: Int arm-type P1, P2, P′1, P′2, and P′3 (green); integration host factor (IHF), H1, H2, and H′ (gray); Xis, X1, X1.5, and X2 (gold); and Fis (pink). The four core-type Int binding sites, C, C′, B, and B′ (blue boxes) are each bound in a C-clamp fashion by the CB and CAT domains, referred to here as the CTD. This is where Int executes isoenergetic DNA strand cleavages and ligations via a high-energy covalent 3′-phospho-tyrosine intermediate. The CTD of Int and the tetrameric Int complex surrounding the two overlap regions are functionally and structurally similar to the Cre, Flp, and XerC/D proteins. Reprinted with permission from reference 36. doi:10.1128/microbiolspec.MDNA3-0051-2014.f1
