FIGURE 8.
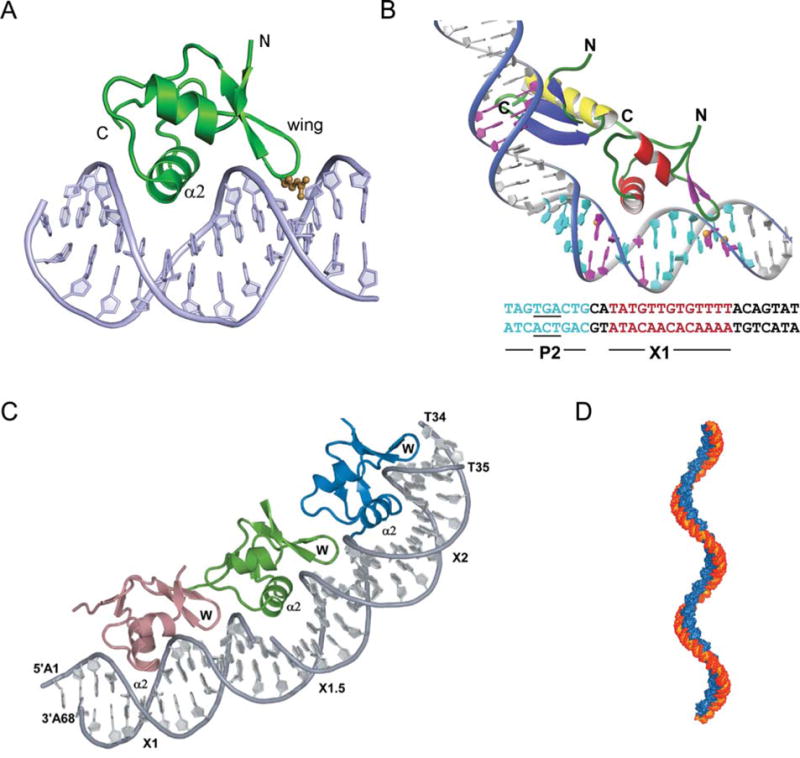
Complex of Xis with DNA. (A) The structure of 1–55XisC28S specifically bound to X2 DNA penetrates adjacent grooves of the duplex by fastening on the phosphodiester backbone. The major groove is filled primarily with helix α2 with the side chains of Glu19, Arg23, and Arg26 playing a major role in specific DNA recognition. The adjacent minor groove is contacted by the “wing” which does not contribute significantly to the specificity of complex formation but does contribute to binding affinity, although to a smaller extent than helix α2. The side-chain of Arg39 (brown) extends along the floor of the minor groove where it makes direct and water-mediated hydrogen bonds. (B) A model for the Int (NTD)-Xis-DNA ternary complex. The Int (NTD) is modeled to interact with the TGA trinucleotide (underlined) of the P2 site (blue) in the DNA major groove. Xis is modeled on the X1 site (magenta) in the same manner as observed in the complex with the X2 site. The C-terminal tail of Xis, which is disordered in solution (not shown), is located adjacent to the C-terminal helix of the NTD of Int to make a protein–protein interaction as shown by mutagenesis and NMR titration data (179). (C) X-ray crystal structure of Xis bound to the Xis binding region reveals the structural basis of cooperative binding. Xis monomers bound to the X1, X1.5, and X2 sites are colored dark salmon, green, and blue, respectively. (D) Structure-based model of an extended Xis-DNA filament. Units of the Xis-DNAX1–X2 crystal structure were stacked end-to-end by superimposing site X1 over X1.5 to assemble a pseudocontinuous helix with a pitch of ~22 nm. Proteins are blue; DNA is orange. Reprinted with permission from reference 139 (A and B) and reference 140 (C and D). doi:10.1128/microbiolspec.MDNA3-0051-2014.f8
