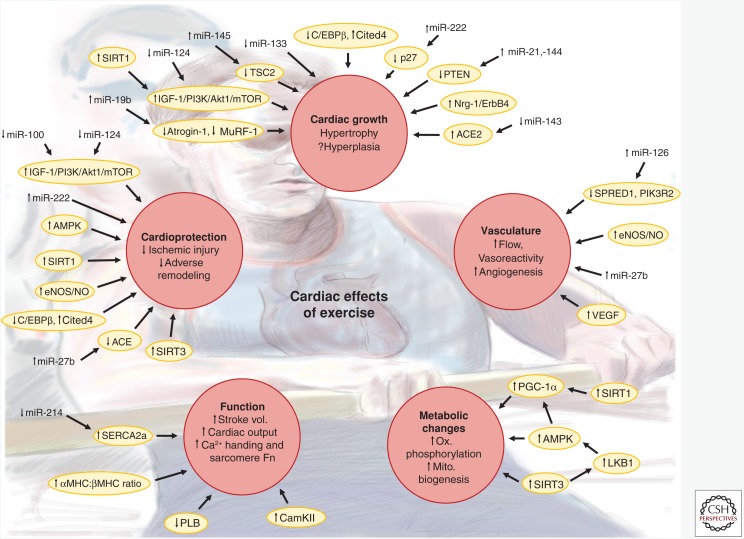
Figure 1.
Cardiac phenotypes induced by exercise. Schematic illustrating the cardiac effects induced by chronic endurance exercise training. Red circles indicate major phenotypes or groups of phenotypes altered in response to exercise training. Yellow ovals indicate key proteins or pathways thought to contribute to the phenotype changes. Vertical arrows next to phenotypes, proteins/pathways, or microRNAs (miRNAs) indicate reported increases or decreases in response to exercise. For proteins/pathways, changes may be either in level or activity. Arrows connecting miRNAs, proteins/pathways, and phenotypes indicate regulation either demonstrated in animal models of exercise or inferred from in vitro experiments and/or in vivo experiments of related contexts. Stroke vol., Stroke volume; Sarcomere Fn, sarcomere function; Ox. phosphorylation, oxidative phosphorylation; Mito. biogenesis, mitochondrial biogenesis; ACE, angiotensin I converting enzyme; ACE2, angiotensin I converting enzyme 2; Akt1, AKT serine/threonine kinase 1; AMPK, AMP-activated kinase; CaMKII, calcium/calmodulin-dependent protein kinase II; C/EBPβ, CCAAT/enhancer binding protein β; Cited4, Cbp/p300 interacting transactivator with Glu/Asp rich carboxy-terminal domain 4; eNOS, endothelial nitric oxide synthase; ErbB4, erb-b2 receptor tyrosine kinase 4; IGF-1, insulin-like growth factor 1; LKB1, serine/threonine liver kinase B1; MHC, myosin heavy chain; mTOR, mechanistic target of rapamycin; MuRF-1, muscle-specific RING finger protein 1; NO; nitric oxide; Nrg-1, neuregulin 1; p27, cyclin-dependent kinase inhibitor 1B (p27); PGC-1α, peroxisome proliferator-activated receptor γ coactivator 1α; PI3K, phosphoinositide-3-kinase; PIK3R2, phosphoinositide-3-kinase regulatory subunit 2; PLB, phospholamban; PTEN, phosphatase and tensin homolog; SERCA2a, sarcoplasmic reticulum calcium ATPase 2 isoform a; SIRT1, sirtuin 1; SIRT3, sirtuin 3; SPRED1, sprouty-related EVH1 domain-containing 1; TSC2, tuberous sclerosis 2; VEGF, vascular endothelial growth factor.