This case series provides detailed clinical, electrophysiologic, structural, and molecular genetic findings in nonsyndromic inherited retinal degenerations associated with CLN3 mutations.
Key Points
Question
What are the clinical and molecular genetic findings of patients with CLN3-associated isolated retinal degeneration compared with retinal disease in juvenile neuronal ceroid lipofuscinosis classically associated with CLN3 mutations?
Findings
This case series of 10 patients with CLN3-associated isolated retinal degeneration identified phenotypic and genotypic differences compared with retinal disease in classic CLN3-associated juvenile neuronal ceroid lipofuscinosis.
Meaning
The findings of this case series suggest that mutations in CLN3 can present with an isolated retinal degeneration that has a later onset than juvenile neuronal ceroid lipofuscinosis, indicating a potential therapeutic window for these patients.
Abstract
Importance
Mutations in genes traditionally associated with syndromic retinal disease are increasingly found to cause nonsyndromic inherited retinal degenerations. Mutations in CLN3 are classically associated with juvenile neuronal ceroid lipofuscinosis, a rare neurodegenerative disease with early retinal degeneration and progressive neurologic deterioration, but have recently also been identified in patients with nonsyndromic inherited retinal degenerations. To our knowledge, detailed clinical characterization of such cases has yet to be reported.
Objective
To provide detailed clinical, electrophysiologic, structural, and molecular genetic findings in nonsyndromic inherited retinal degenerations associated with CLN3 mutations.
Design, Setting, and Participants
A multi-institutional case series of 10 patients who presented with isolated nonsyndromic retinal disease and mutations in CLN3. Patient ages ranged from 16 to 70 years; duration of follow-up ranged from 3 to 29 years.
Main Outcomes and Measures
Longitudinal clinical evaluation, including full ophthalmic examination, multimodal retinal imaging, perimetry, and electrophysiology. Molecular analyses were performed using whole-genome sequencing or whole-exome sequencing. Electron microscopy studies of peripheral lymphocytes and CLN3 transcript analysis with polymerase chain reaction amplification were performed in a subset of patients.
Results
There were 7 females and 3 males in this case series, with a mean (range) age at last review of 37.1 (16-70) years. Of the 10 patients, 4 had a progressive late-onset rod-cone dystrophy, with a mean (range) age at onset of 29.7 (20-40) years, and 6 had an earlier onset rod-cone dystrophy, with a mean (range) age at onset of 12.1 (7-17) years. Ophthalmoscopic examination features included macular edema, mild intraretinal pigment migration, and widespread atrophy in advanced disease. Optical coherence tomography imaging demonstrated significant photoreceptor loss except in patients with late-onset disease who had a focal preservation of the ellipsoid zone and outer nuclear layer in the fovea. Electroretinography revealed a rod-cone pattern of dysfunction in 6 patients and were completely undetectable in 2 patients. Six novel CLN3 variants were identified in molecular analyses.
Conclusions and Relevance
This report describes detailed clinical, imaging, and genetic features of CLN3-associated nonsyndromic retinal degeneration. The age at onset and natural progression of retinal disease differs greatly between syndromic and nonsyndromic CLN3 disease, which may be associated with genotypic differences.
Introduction
Juvenile neuronal ceroid lipofuscinosis (JNCL), or Batten disease, is one form of a group of progressive neurodegenerative diseases characterized by lysosomal accumulation of ceroid lipopigments. Patients with JNCL experience retinal degeneration, seizures, severe motor and cognitive deterioration, and shortened life expectancy. Clinically, this group of disorders is classified by age at disease onset, with genetic heterogeneity with 14 causative genes identified to date (CLN1 through CLN14).
Juvenile neuronal ceroid lipofuscinosis is the most prevalent form of the NCL disorders, with an incidence of 1 in 25 000 births. It is associated with biallelic mutations in CLN3, with a common founder 1.02-kb deletion mutation occurring in homozygosity in 76% of patients and compound heterozygosity in an additional 22% of patients. Vision loss is usually the presenting symptom, occurring between 4 and 8 years of age, with rapid progression to legal blindness within 2 to 4 years. Ophthalmic findings include optic nerve pallor, bull’s-eye maculopathy, intraretinal pigmentation, and severely abnormal findings on electroretinography. Neurological symptoms subsequently occur, beginning with speech disturbances and cognitive decline at 8 to 9 years, seizures at 5 to 18 years, and extrapyramidal and motor decline at 12 to 20 years, leading to death in the second decade of life.
With the growing use of next-generation sequencing in inherited retinal degenerations (IRDs), mutations in genes previously associated with syndromic retinal degenerations are increasingly being identified in patients with nonsyndromic IRD. Genes previously associated with syndromic diseases, such as Bardet-Biedl, Usher, Senior-Loken, Mainzer-Saldino, and Joubert syndromes, mucopolysaccharidosis, and neuronal ceroid lipofuscinosis have been reported in patients with isolated retinal disease. Mutations in CLN3 were recently identified in patients with nonsyndromic IRD in 2 large-scale molecular diagnostic studies, but there was a lack of phenotypic information. To our knowledge, the present report is the first detailed phenotypic characterization of CLN3-associated nonsyndromic retinal disease.
Methods
Participants
The study followed the tenets of the Declaration of Helsinki and was approved by the Oregon Health & Science University Institutional Review Board, the research management committee of Moorfields Eye Hospital in London, England, and the research ethics board of the Hospital for Sick Children in Toronto, Canada. Patients with mutations in CLN3 were enrolled from the inherited retinal disease clinics at Moorfields Eye Hospital (MEH; patients MEH1 to MEH6), the Hospital for Sick Children (SK; patients SK1 to SK3), and the Casey Eye Institute of the Oregon Health & Science University (CEI; patient CEI1). Written informed consent was obtained from all participants prior to study inclusion.
Retinal Imaging
Each participant underwent full clinical examination. Color fundus photography was conducted using 35° (Topcon; Carl Zeiss) or 60° (Canon USA) color fundus photography or ultra–wide-field confocal scanning laser imaging (Optos). Fundus autofluorescence was performed with 30° (Spectralis), 55° (Spectralis), or ultra–wide-field (Optos) imaging with excitation wavelength 488 nm (Spectralis) and 532 nm (Optos). Spectral-domain optical coherence tomography scans (Spectralis; Cirrus) and kinetic visual fields (Goldmann or Ocotopus 900 Perimeter; Haag-Streit) were performed.
Electrophysiology
Full-field electroretinography (ERG) and multifocal ERG were performed using Burian-Allen (at CEI and SK), DTL (SK), or gold foil (at MEH) electrodes and pattern ERG with gold foil (at MEH) recording electrodes under protocols that conformed to the International Society for Clinical Electrophysiology of Vision standards.
Molecular Analysis
Briefly, genomic DNA was isolated from peripheral blood. Whole-exome sequencing analysis was performed on patients MEH1 and MEH6 at AROS Applied Biotechnology using SureSelectXT Human All Exon version 5 exon capture (Agilent Technologies) followed by sequencing on a HiSeq2000 platform (Illumina), as previously described. Whole-genome sequencing on patients MEH2-MEH5 was performed as part of a large collaborative study in the National Institute for Health Research BioResource: Rare Diseases Project, Specialist Pathology Evaluating Exomes in Diagnostics using a HiSeq2500 platform (Illumina). APEX microarray screening (Asper Biotech) was performed on patient MEH6 using a genotyping microarray containing more than 700 disease-causing variants for 28 retinal dystrophy genes, as previously described. Molecular analysis for patients CEI1 and SK1-3 was performed using a custom-designed next-generation sequencing panel containing more than 3500 target sequences in 183 genes (rd v4 panel).
Variants were annotated using RefSeq gene sequence number NM_000086.2, Ensembl gene number ENSG00000188603, transcript number ENST00000360019, and protein number ENSP00000353116, as appropriate. Novel disease-causing variants were identified as such when not previously associated with Batten disease or nonsyndromic IRDs in the literature or the online Neuronal Ceroid Lipofuscinosis Mutation and Patient Database (http://www.ucl.ac.uk/ncl/CLN3mutationtable.htm). The pathogenicity of missense mutations was predicted by PolyPhen2 and intronic splice variants by the Berkeley Drosophila Genomic Project (Splice Site Prediction by Neural Network; http://fruitfly.org). Variants were also noted to be novel when absent or low in allele frequency in databases from the Broad Institute (gnomAD browser), the NCBI dbSNP, and the Exome Aggregation Consortium, with allele frequencies reported from the Broad Institute. Segregation analysis was performed in available family members for 6 patients with results consistent with biallelic mutations (patients MEH3, MEH6, CEI1, and SK1-3) (eFigure 1 in the Supplement).
Additional Testing
Electron microscopy studies were performed on peripheral lymphocytes in 3 patients. Briefly, blood samples were centrifuged to obtain the buffy coat and fixed in 1.5% glutaraldehyde, 1.5% paraformaldehyde in 0.1M sodium cacodylate buffer pH 7.4 with 0.05M sucrose and 0.25 calcium chloride overnight. Electron microscopy images were obtained on a transmission electron microscope interfaced to a camera. Lymphoblasts from the patient and controls were cultured, with RNA extraction, reverse-transcription, and quantitative reverse-transcription polymerase chain reaction performed, as previously described. Additionally, immunocytochemistry measuring autofluorescence levels was performed on the cultured lymphoblasts with DAPI (4′,6-diamidino-2-phenylindole) staining.
Results
Ophthalmic evaluations were conducted on 10 patients from 9 families (3 male, 7 female) last seen at ages ranging from 16 to 70 years (mean, 37.1 years), with key clinical features summarized in Table. Age at symptom onset ranged from the first to the fourth decade of life (mean age, 23 years). Best-corrected visual acuity ranged from −0.1 logMAR (Snellen acuity, 20/16) to light perception. Four patients showed a late-onset phenotype, with visual symptoms beginning in the second to fourth decade of life (MEH1-4) and relatively preserved visual acuity of +0.2 logMAR (Snellen acuity, 20/32) or better at last follow-up. In contrast, 6 patients presented with early-disease onset before the second decade of life and significantly decreased visual acuity within the first decade of disease onset (CEI1, MEH5-6, and SK1-3).
Table. Clinical Summary of 10 Patients.
| Characteristic | Patient ID (Institutional Identifier) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MEH1 (GC18875) | MEH2 (GC20465) | MEH3 (GC20390) | MEH4 (GC18983) | CEI1 (CEI26198) | SK1 | SK2 | MEH5 (GC20101) | MEH6 (GC3513) | SK3 | |
| Age at last visit, y/sex | 36/F | 30/M | 54/F | 51/M | 24/F | 21/F | 23/F | 16/F | 49/F | 70/M |
| Age at onset, y | 30s | 20s | Late 40s | 30s | Late teens | <10 | Early teens | Early teens | <10 | Early teens |
| Presenting symptom | ||||||||||
| VA loss | Absent | Absent | Absent | Absent | Mild | Mild | Mild | Mild | Mild | Mild |
| Nyctalopia | Mild | Mild | Mild | Mild | Mild | Mild | Mild | Mild | Mild | Mild |
| BCVA snellen | 20/16 | 20/30 | 20/20 | 20/32 | 20/125 | 20/80 | 20/125 | NA | HM | LP |
| Fundus findings | ||||||||||
| Mac atrophy | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Absent | Moderate | Moderate |
| Bone spicules | Absent | Absent | Absent | Mild | Intermittent | Moderate | Intermittent | Absent | Absent | Moderate |
| AF | ||||||||||
| Central hypoAF | Moderate | Mild | Mild | Moderate | Moderate | Mild | Mild | Mild | Severe | Absent |
| HyperAF ring | Mild | Mild | Absent | Mild | Moderate | Mild | Mild | Intermittent | Absent | Absent |
| Periph hypoAF | Severe | Moderate | Mild | Severe | Intermittent | Mild | Moderate | Moderate | Severe | Severe |
| OCT | ||||||||||
| Fov | ||||||||||
| EZ loss | Absent | Intermittent | Mild | Mild | Severe | Moderate | Moderate | Severe | Severe | Severe |
| ONL loss | Intermittent | Mild | Mild | Moderate | Moderate | Moderate | Moderate | Moderate | Severe | Severe |
| PF | ||||||||||
| EZ loss | Severe | Severe | Moderate | Severe | Severe | Intermittent | Mild | Severe | Severe | Severe |
| ONL loss | Severe | Severe | Severe | Moderate | Moderate | Severe | Severe | Moderate | Severe | Severe |
| Mac edema | Absent | Moderate | Severe | Mild | Absent | Severe | Absent | Absent | Absent | Absent |
| VF (age, y) | ||||||||||
| Central scotoma | Absent | Absent | Mild | Absent | Mild | Absent | Absent | NA | Severe | Absent |
| VF constriction | To 30° (30) | Patchy loss (27) | To 10° (54) | To 5° (51) | N/I/S (23) | N/I/S (21) | To 30° (23) | NA | ND (47) | To 10° (61) |
| Electrophysiology (age, y) | ||||||||||
| ffERG: rod, cone function ↓ | Rod: ↓↓ Cone: ↓ (30) |
NA | Rod: ND Cone: ↓ (51) |
Rod: ↓↓↓ Cone: ↓ (37) Rod: ND Cone: ↓↓ (42) |
Rod: ↓↓↓ Cone: ↓↓ (23) |
Rod: ND Cone: ↓↓↓ (10) |
Rod: ND Cone: ND (12) |
Rod: ND Cone: ↓↓↓ (16) |
NA | Rod: ND Cone: ND (41) |
| PERG | ↓↓ | NA | ↓↓ | ↓↓ | NA | NA | NA | ↓↓↓ | NA | NA |
| mfERG: paracentral, Central | PC: ↓ C: Spared |
NA | PC: ↓ C: Spared |
PC: ↓↓ C: Spared |
NA | NA | NA | NA | NA | NA |
| Genetic analysis | ||||||||||
| Allele 1 | c.1213C>T (R405W) | 1-kb del | 1-kb del | c.1213C>T (R405W) | 1-kb del | c.883G>T (E295X) | c.883G>T (E295X) | 1-kb del | 1-kb del | 1-kb del |
| Allele 2 | c.1213C>T (R405W) | c.988G>A (V330I)a | c.837+5G>A (splice site)a | c.1213C>T (R405W) | c.461-3C>G (splice site)a | c.917T>A (L306H)a | c.917T>A (L306H)a | c.853A>G (I285V)a | c.1213C>T (R405W) | c.375-3C>G (splice site)a |
Abbreviations: AF, autofluorescence; BCVA, best-corrected visual acuity; C, central; c, cDNA sequence with nucleotide position and change; CEI, Casey Eye Institute; ERG, electroretinography; EZ, ellipsoid zone; ffERG, full-field ERG; Fov, foveal; hyperAF, hyper-autofluorescence; hypoAF, hypo-autofluorescence; LP, light perception; MEH, Moorfields Eye Hospital; mfERG, multifocal ERG; NA, not applicable; ND, not detectable; N/I/S, nasal/inferior/superior; OCT, optical coherence tomography; ONL, outer nuclear layer; mac, macular; PC, paracentral; PERG, pattern ERG; periph, peripheral; PF, parafoveal; SK, the Hospital for Sick Children; VA, visual acuity; VF, visual field; 1-kb del, c.461-280_677+382del966 (p.Gly154Alafs*29,Val155_Gly264del); ↓, decreased.
Novel mutation.
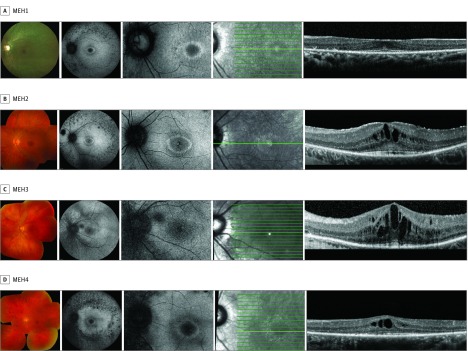
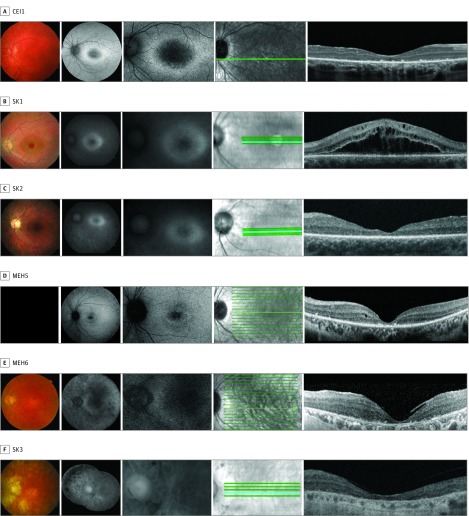
Ophthalmoscopic findings included macular edema, mild intraretinal bone spicule migration, and significant macular atrophy. Macular edema was present in 4 patients (MEH2-4 and SK1) (Figure 1 and Figure 2). Edema was unresponsive to topical dorzolamide but partially responsive to brinzolamide in 1 patient (MEH3). Sparse intraretinal bone spicule-like pigment migration was present in 2 patients (CEI1 and SK2) and mild to moderate in 4 patients (MEH4 and SK1-3) (Figure 1 and Figure 2). Macular atrophy with retinal pigment epithelium loss was evident in the 2 patients with the worst visual acuity (MEH6 and SK3).
Figure 1. Composite of Retinal Imaging From the Left Eyes of Patients With Late-Onset CLN3-Associated Rod-Cone Dystrophy.
Imaging from each patient includes color fundus photograph (first panel), fundus autofluorescence (second and third panels), and en face infrared image (fourth panel) demarcating the horizontal optical coherence tomography scan line (fifth panel).
Figure 2. Composite of Retinal Imaging From the Left Eyes of Patients With Early-Onset CLN3-Associated Rod-Cone Dystrophy.
Imaging from each patient includes color fundus photograph (first panel), fundus autofluorescence (second and third panels), and en face infrared image (fourth panel) demarcating the horizontal optical coherence tomography scan line (fifth panel). No color fundus photograph was able to be obtained for patient MEH5 (first panel).
Fundus autofluorescence imaging showed central retinal hypoautofluorescence beyond the normal macular pigment hypoautofluorescence, with a surrounding hyperautofluorescent ring in 7 patients (CEI1, MEH1-2, MEH4-5, and SK1-2) (Figure 1 and Figure 2). There were also patchy hypoautofluorescence in the midperiphery and along the arcades, ranging from mild (CEI1 and MEH3) or moderate (MEH2 and MEH5) to dense (MEH1 and MEH4). Two patients with significant visual acuity loss to hand motion and light perception showed generalized hypoautofluorescence (MEH6 and SK3).
Optical coherence tomography imaging in 4 patients with preserved visual acuity (MEH1-4) showed a relatively intact ellipsoid zone at the fovea, despite presence of macular edema (MEH2-3). However, outside of the focal foveal sparing, the ellipsoid zone was absent and outer nuclear layer significantly diminished. Additionally, 2 of 4 patients showed an area of hyperreflectance above the external limiting membrane at the fovea (MEH1 and MEH2), suggestive of nonspecific debris. Six patients with decreased visual acuity showed moderate to complete foveal outer nuclear layer and ellipsoid zone loss (CEI1, MEH5-6, and SK1-3). Hyperreflective deposits at the level of the retinal pigment epithelium were present in some patients with advanced disease (CEI1 and MEH5-6).
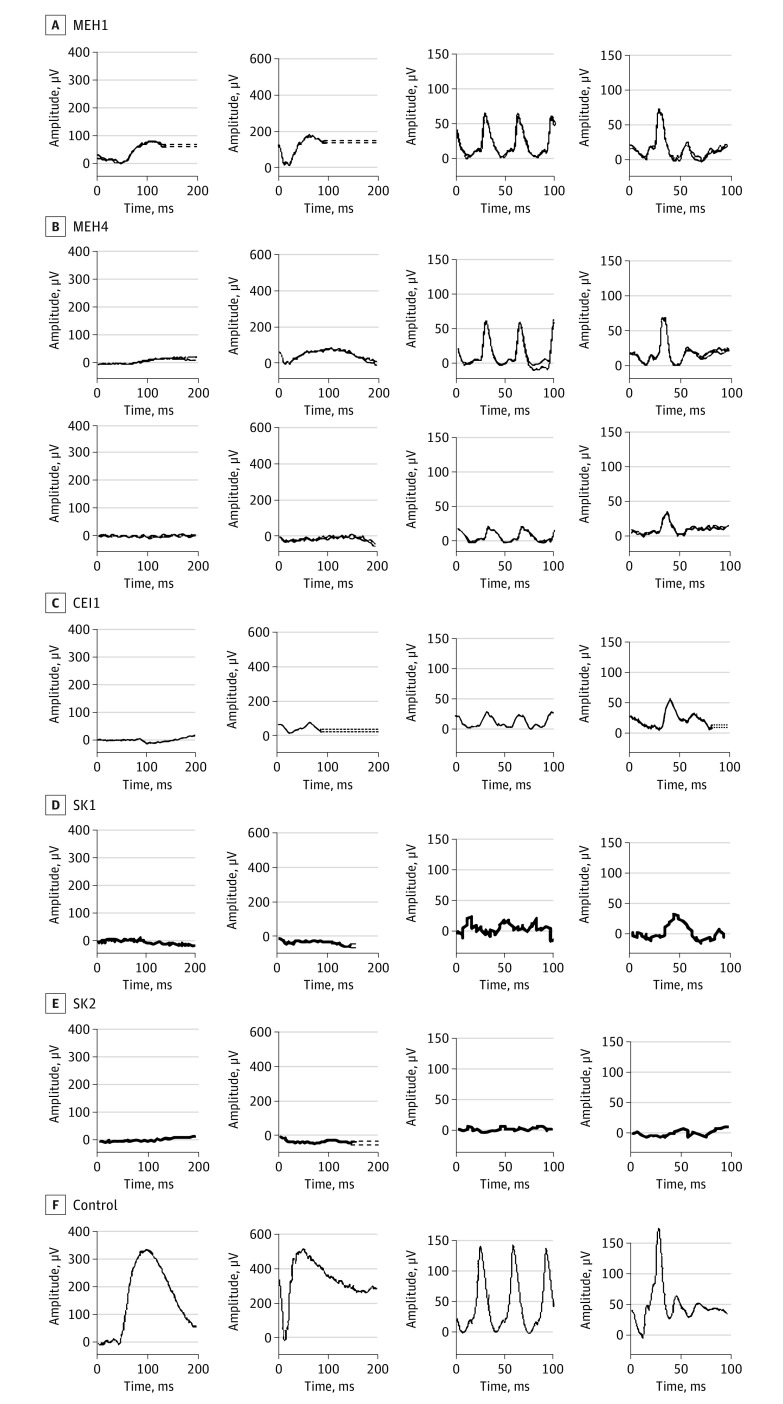
Electroretinography was performed in 8 of 10 patients (CEI1, MEH1, MEH3-5, and SK1-3) with all patients showing a rod-cone dystrophy. All patients showed markedly abnormal scotopic responses shortly after disease onset, with variable abnormalities in photopic ERGs (Figure 3) (Table) (eFigure 2 in the Supplement). Patients with late-onset disease demonstrated relatively preserved photopic responses (MEH1, MEH3, and MEH4), although longitudinal ERGs showed progressive decline (CEI1 and MEH4) (Table) (Figure 3). There was marked paracentral loss of cone function with some central sparing on multifocal ERG (MEH1, MEH3, and MEH4) (eFigure 3 in the Supplement). Patients with earlier onset of disease showed severe loss of both rod and cone responses (SK1 and SK2) (Table) (Figure 3).
Figure 3. Full-Field Electroretinograms.
Results of full-field electroretinograms under dim scotopic (dark-adapted [DA] 0.01; first panel), bright scotopic (DA 10.0; second panel), photopic flicker (light-adapted [LA] 3.0, 30 Hz; third panel), and photopic single flash (LA 3.0; fourth panel) intensities. Results for patient MEH4 shown from testing performed in his late 30s and 5 years later.
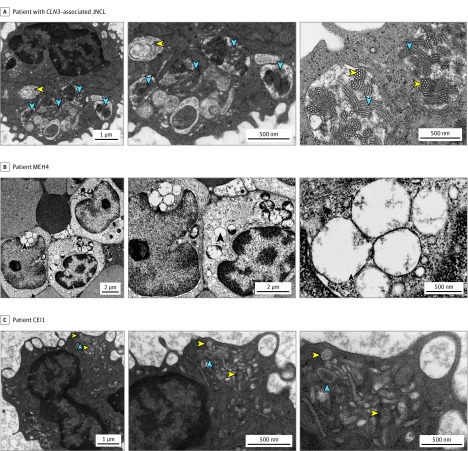
None of the 10 patients had signs or symptoms of neurologic involvement. Head imaging was performed in 2 patients (MEH1 and MEH4), with an incidental fascinoma or capillary telangiectatic lesion observed on brain magnetic resonance imaging in MEH4. Electron microscopy of peripheral lymphocytes was performed in 3 patients (CEI1, MEH1, and MEH4). Classic findings of prominent fingerprint profiles and vacuolation resulting from lipofuscin accumulation and defective vacuolar homeostasis in JNCL were not observed, although MEH4 had very intermittent large empty vacuoles (Figure 4).
Figure 4. Electron Microscopy (EM) of Peripheral Lymphocytes.
A, A patient with CLN3-associated juvenile neuronal ceroid lipofuscinosis (JNCL). Left and middle panels, Classic EM findings in a patient with CLN3-associated JNCL with foamy vacuoles (yellow arrowheads) and lysosomal storage material forming fingerprint profile structures (blue arrowheads). Right panel, Fingerprint profiles in sagittal (blue arrowheads) and coronal cuts (yellow arrowheads), observed at a high magnification. B, Patient MEH4. The classic EM findings associated with CLN3-associated JNCL were not observed in patient MEH4, who had unremarkable lymphocytes except for the occasional large empty vacuoles (arrowheads). C, Patient CEI1. Classic JNCL findings were also absent in patient CEI1, who had unremarkable-appearing lysosomes (yellow arrowheads) and occasional vacuoles (blue arrowheads) without accumulation of storage material.
Variants in CLN3 were identified in all 10 patients, including 2 patients with homozygous and 8 patients with compound heterozygous variants. Six novel CLN3 variants—3 missense and 3 intronic—were identified (Table) (eTable in the Supplement). All 3 novel missense variants (I285V, V330I, and L306H) occurred as compound heterozygous mutations (Table). Population allele frequencies are very low at 3.96 × 10−6 for I285V and 3.21 × 10−5 for V330I; to our knowledge, L306H has not previously been reported as a population variant (gnomAD browser; Broad Institute). Three novel intronic variants (c.375-3C>G, c.461-3C>G, and c.837+5G>A) were identified, all occurring as a compound heterozygous variant with the 1.02-kb deletion. While resultant altered splice transcripts remain unknown, all 3 intronic variants fall within predicted splice donor or acceptor sites (Berkeley Drosophila Genome Project) (eTable in the Supplement) and may therefore lead to altered transcription. Decreased transcription levels were confirmed with c.461-3C>G (CEI1) (eFigure 4 in the Supplement). Population variances of intronic variants were very low, with allele frequency of 4.29 × 10−6 for c.461-3C>G and 6.34 × 10−5 for c.837+5G>A; to our knowledge, c.375-3C>G has never been previously reported as a population variant.
Discussion
Mutations in CLN3 are classically associated with JNCL, a neurodegenerative disease with early retinal degeneration in the first decade of life followed by progressive neurological deterioration and death in the second to third decade. CLN3 mutations have recently been molecularly identified in cases of nonsyndromic isolated retinal degeneration, but a description of the clinical phenotype and natural history was lacking.
Patients in this study fell into 2 distinct phenotypic groups: a late-onset and an early-onset retinal phenotype. Four patients showed a late-onset phenotype with visual symptoms beginning in the second to fourth decade of life (MEH1-4). Central vision was maintained in the first decade of disease followed by slowly progressive decline in the second to third decade after disease onset. In contrast, 6 patients presented with earlier-onset retinal dysfunction before the second decade of life (CEI1, MEH5-6, and SK1-3); onset of visual loss in these patients was still later than in patients with JNCL, which has a mean onset of 4 to 5.4 years.
All patients demonstrated rod-cone dystrophy with marked diminished rod function and significant but variable cone system dysfunction. Both dark-adapted and light-adapted ERG responses were minimal in patients with early-onset disease, even at the youngest age (SK1-2). Patients with late-onset disease demonstrated residual dark-adapted responses with a reduced b-wave to a-wave ratio and a profoundly subnormal a-wave amplitude, likely reflecting responses arising from dark-adapted cones in the absence of rod function (MEH1, MEH4, and CEI1) (Figure 3) (eFigures 2 and 5 in the Supplement). Light-adapted ERG responses were present but decreased and continued to progressively decrease in MEH4. Therefore, the delayed and progressive decline in cone function may benefit from therapeutic intervention, such as AAV-mediated CLN3 gene replacement, which was recently demonstrated in the rodent retina.
CLN3-associated isolated retinal degeneration shows greater macular involvement than the rod-cone degeneration of retinitis pigmentosa. While there was significant macular disease, focal preservation of foveal cone structure and function was observed on optical coherence tomography, multifocal ERG, and visual fields in 4 patients (MEH1-4). Additionally, 4 of 10 patients demonstrated cystoid macular edema, occurring more prevalently than in retinitis pigmentosa and not commonly found in CLN3-associated JNCL. Three of these 4 patients had late-onset disease, suggesting cystoid macular edema as an early clinical feature that progress to macular atrophy. A central hypoautofluorescence with a hyperautofluorescent ring and patchy midperipheral hypofluorescence in a mottled pattern was observed in the majority of the late-onset patients. This paracentral ring lesion is common in retinitis pigmentosa and may reflect accumulation of lipofuscin at the transition zone between unhealthy paracentral and healthy central cones encircling areas of preserved photopic function. Patients with advanced disease demonstrated diffuse hypoautofluorescence (MEH6 and SK3), reflecting severe retinal pigment epithelium and retinal atrophy. Even in advanced disease, characteristic retinitis pigmentosa intraretinal pigment migration was not a prominent finding in our patients.
The spectrum of CLN3-associated disease ranges from JNCL, protracted JNCL with classic early retinal degeneration with delayed neurologic onset, and nonsyndromic retinal degeneration without neurologic involvement. Although a protracted JNCL variant has been described, patients in this cohort remained without neurological symptoms well beyond this age. The importance of CLN3 in retinal cell function is clear by its association with isolated retinal degeneration, invariably early visual dysfunction in classic and protracted JNCL, and subclinical visual changes in carriers of the CLN3 1.02-kb deletion mutation.
While its exact cellular function remains unknown, CLN3 belongs to the major facilitator superfamily of membrane transporters and has been implicated in the transport of protons, amino acids, neurotransmitters, and proteins in lysosomes, at the Golgi, and across synapses. Electron microscopy studies localizes CLN3 in the retina to photoreceptor inner segments, inner retinal cells, and Muller glia. Mouse and in vitro studies have implicated CLN3 in trafficking and stability of the inner-segment protein Na+/K+-ATPase. Neuronal ceroid lipofuscinosis proteins may also regulate palmitoylation, an important posttranslational lipid modification for protein trafficking into outer segments. Lastly, disruption of essential outer segment phagocytosis with defective phagosome-lysosomal fusion has been observed.
CLN3 is a 438–amino acid, 13-exon, transmembrane protein that contains 6 transmembrane domains, an amphipathic helical domain, and 2 lysosomal targeting motifs (EEEX8LI and MX9G) that traffic glycosylated CLN3 from the Golgi to lysosomes (Figure 5). A 1.02-kb deletion in CLN3 is a common founder mutation present in 96% of patients with JCNL; 74% are homozygous and 22% are heterozygous for this mutation. The major predicted protein product from this deletion consists of the first 153 amino acids from exons 1 to 6 plus an additional 28 novel amino acids. Although this mutation results in decreased mRNA transcript levels (eFigure 4 in the Supplement), it remains controversial whether alternatively spliced products are expressed or degraded. Detailed in silico analysis predicts nonsense-mediated decay leading to loss of function with homozygous 1.02-kb deletions. However, in vitro functional assays in cultured fibroblasts and fission yeast suggest that, if expressed, the resulting protein product partially rescues abnormal enlargement of lysosomes occurring with complete loss of CLN3.
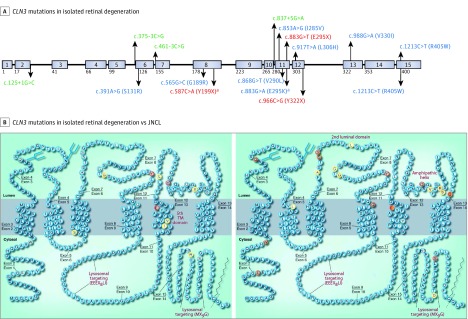
Figure 5. CLN3 Mutations Associated With Isolated Retinal Degeneration.
A, CLN3 mutations observed in isolated retinal degeneration in the current series of 10 patients (above) and in the literature (below). Intronic mutations are indicated in green, missense mutations in blue, and truncation mutations in red. Mutations associated with isolated retinal degeneration observed in this series. B, CLN3 mutations associated with isolated retinal degeneration (left panel) compared with juvenile neuronal ceroid lipofuscinosis (JNCL; right panel). Multiple mutations associated with isolated retinal degenerations (left) are in the fifth transmembrane domain compared with more mutations associated with JNCL (right) in the luminal loops. Adapted with permission from Cotman et al (Taylor & Francis Ltd; www.tandfonline.com).
aMutations also associated with patients with CLN3-associated JNCL with neurological symptoms.
Although less prevalent, evaluation of compound heterozygous mutations occurring with the 1.02-kb deletion may provide insights on genotype-phenotype correlations. The 1.02-kb deletion mutation leads to complete loss of exons 7 and 8 that form the second luminal loop of CLN3. Interestingly, compound mutations associated with JNCL are also clustered at this site (ie, S161X, S162X, A158P, L170P, G187R, and G189A), suggesting the presence of critical substrate binding sites there (Figure 5). Rare cases of homozygous mutations in the second luminal loop show a less severe neurologic phenotype than the 1.02-kb deletion. For example, G189A is associated with JNCL when paired with the 1.02-kb deletion, but homozygous G189R mutations are associated with isolated retinal degeneration. Homozygous mutations in Y199X (eTable in the Supplement) associated with isolated retinal degeneration and protracted JNCL may show lower neurologic severity owing to the additional 46 amino acids from exons 7 and 8 and lower likelihood of nonsense-mediated decay compared with homozygous 1.02-kb deletions.
We observed that compound heterozygous mutations associated with isolated retinal degeneration or protracted JNCL were clustered in or near the fifth transmembrane domain, with 4 missense variants in this region, 3 novel to this study (I285V, V290L, and L306H) (Figure 5). A specific missense R405W mutation, located in a conserved region upstream of the C-terminal lysosomal targeting domain (MX9G), has been solely associated with nonsyndromic retinal degeneration. It was previously identified in 2 autosomal recessive retinitis pigmentosa probands as a homozygous mutation and as a compound heterozygous mutation with an intronic variant (eTable in the Supplement). In the present series, it occurs as a homozygous mutation (MEH1 and MEH4) and compound heterozygous mutation with the 1.02-kb deletion (MEH6) (Table) (Figure 5) (eTable in the Supplement). Patients with homozygous R405W mutations show late-onset compared with the early-onset phenotype of MEH6, further suggesting that the R405W mutation is less deleterious than the 1.02-kb deletion. Evaluation of the 1.02-kb deletion compound heterozygous mutations associated with nonsyndromic retinal degeneration suggests that the R405W (MEH6) and I285V (MEH5) mutations cause a more severe retinal phenotype than the V330I (MEH2) mutation.
Altogether, the data confirm that the common founder 1.02-kb deletion mutation likely leads to the loss of an important CLN3 function affecting both the central nervous system and retinal neurons owing to loss of exons 7 and 8 that make up the second luminal loop; this deleterious effect is also observed in compound heterozygous mutations in the luminal loops. Mutations affecting the fifth transmembrane and the C-terminal lysosomal targeting domain appear to be associated with milder retinal dysfunction and decreased or absent neurologic involvement. Newly identified variants and proposed genotype-phenotype observations will continue to be confirmed and refined with further identification of CLN3 mutations in patients with isolated retinal degeneration.
Limitations
Our study had limitations. Owing to the retrospective nature of the case series, clinical studies may have been unavailable for some patients. Parental DNA samples were unavailable for segregation analysis for some of the families described in this study. Electroretinography was unavailable in 3 patients, and specialized studies, such as electron microscopy and magnetic resonance imaging, were available for only several patients.
Conclusions
To our knowledge, this series of patients is the largest to date with isolated retinal dysfunction associated with mutations in CLN3, a gene typically associated with generalized neurological dysfunction. The mechanisms underlying the differences in neurological involvement are yet to be established and underscore the importance of genotype-phenotype studies between classic JNCL, protracted JNCL, and isolated nonsyndromic retinal degeneration. Further detailed clinical studies are also required to evaluate potential genotype-phenotype correlations in the disease spectrum observed in this study.
eTable. Functional prediction of CLN3 mutations associated with isolated retinal degeneration.
eFigure 1. Pedigrees and mutation segregation in 5 families for 6 patients.
eFigure 2. Supplementary full-field electroretinograms.
eFigure 3. Multifocal electroretinography of patients with late-onset retinal degeneration who retain residual full-field cone responses.
eFigure 4. Evaluation of the effect of intronic mutation c.461-3C>G on CLN3 transcript level and autofluorescent phenotype.
eFigure 5. Patients show electroretinography responses with reduced b-wave to a-wave ratio under scotopic conditions.
References
- 1.Zeman W, Dyken P. Neuronal ceroid-lipofuscinosis (Batten’s disease): relationship to amaurotic family idiocy? Pediatrics. 1969;44(4):570-583. [PubMed] [Google Scholar]
- 2.Wisniewski KE, Zhong N, Philippart M. Pheno/genotypic correlations of neuronal ceroid lipofuscinoses. Neurology. 2001;57(4):576-581. [DOI] [PubMed] [Google Scholar]
- 3.Mole SE, Williams RE, Goebel HH. Correlations between genotype, ultrastructural morphology and clinical phenotype in the neuronal ceroid lipofuscinoses. Neurogenetics. 2005;6(3):107-126. [DOI] [PubMed] [Google Scholar]
- 4.Cárcel-Trullols J, Kovács AD, Pearce DA. Cell biology of the NCL proteins: what they do and don’t do. Biochim Biophys Acta. 2015;1852(10, pt B):2242-2255. [DOI] [PubMed] [Google Scholar]
- 5.The International Batten Disease Consortium Isolation of a novel gene underlying Batten disease, CLN3. Cell. 1995;82(6):949-957. [DOI] [PubMed] [Google Scholar]
- 6.Uvebrant P, Hagberg B. Neuronal ceroid lipofuscinoses in Scandinavia: epidemiology and clinical pictures. Neuropediatrics. 1997;28(1):6-8. [DOI] [PubMed] [Google Scholar]
- 7.Bohra LI, Weizer JS, Lee AG, Lewis RA. Vision loss as the presenting sign in juvenile neuronal ceroid lipofuscinosis. J Neuroophthalmol. 2000;20(2):111-115. [DOI] [PubMed] [Google Scholar]
- 8.Spalton DJ, Taylor DS, Sanders MD. Juvenile Batten’s disease: an ophthalmological assessment of 26 patients. Br J Ophthalmol. 1980;64(10):726-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weleber RG. The dystrophic retina in multisystem disorders: the electroretinogram in neuronal ceroid lipofuscinoses. Eye (Lond). 1998;12(pt 3b):580-590. [DOI] [PubMed] [Google Scholar]
- 10.Eksandh LB, Ponjavic VB, Munroe PB, et al. . Full-field ERG in patients with Batten/Spielmeyer-Vogt disease caused by mutations in the CLN3 gene. Ophthalmic Genet. 2000;21(2):69-77. [PubMed] [Google Scholar]
- 11.Williams RE, Aberg L, Autti T, Goebel HH, Kohlschütter A, Lönnqvist T. Diagnosis of the neuronal ceroid lipofuscinoses: an update. Biochim Biophys Acta. 2006;1762(10):865-872. [DOI] [PubMed] [Google Scholar]
- 12.Aldahmesh MA, Safieh LA, Alkuraya H, et al. . Molecular characterization of retinitis pigmentosa in Saudi Arabia. Mol Vis. 2009;15:2464-2469. [PMC free article] [PubMed] [Google Scholar]
- 13.Riazuddin SA, Iqbal M, Wang Y, et al. . A splice-site mutation in a retina-specific exon of BBS8 causes nonsyndromic retinitis pigmentosa. Am J Hum Genet. 2010;86(5):805-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Estrada-Cuzcano A, Koenekoop RK, Senechal A, et al. . BBS1 mutations in a wide spectrum of phenotypes ranging from nonsyndromic retinitis pigmentosa to Bardet-Biedl syndrome. Arch Ophthalmol. 2012;130(11):1425-1432. [DOI] [PubMed] [Google Scholar]
- 15.Méndez-Vidal C, Bravo-Gil N, González-Del Pozo M, et al. . Novel RP1 mutations and a recurrent BBS1 variant explain the co-existence of two distinct retinal phenotypes in the same pedigree. BMC Genet. 2014;15:143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bujakowska KM, Zhang Q, Siemiatkowska AM, et al. . Mutations in IFT172 cause isolated retinal degeneration and Bardet-Biedl syndrome. Hum Mol Genet. 2015;24(1):230-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goyal S, Jäger M, Robinson PN, Vanita V. Confirmation of TTC8 as a disease gene for nonsyndromic autosomal recessive retinitis pigmentosa (RP51) [published online July 21, 2015]. Clin Genet. [DOI] [PubMed] [Google Scholar]
- 18.Shevach E, Ali M, Mizrahi-Meissonnier L, et al. . Association between missense mutations in the BBS2 gene and nonsyndromic retinitis pigmentosa. JAMA Ophthalmol. 2015;133(3):312-318. [DOI] [PubMed] [Google Scholar]
- 19.Khan AO, Decker E, Bachmann N, Bolz HJ, Bergmann C. C8orf37 is mutated in Bardet-Biedl syndrome and constitutes a locus allelic to non-syndromic retinal dystrophies. Ophthalmic Genet. 2016;37(3):290-293. [DOI] [PubMed] [Google Scholar]
- 20.Rivolta C, Sweklo EA, Berson EL, Dryja TP. Missense mutation in the USH2A gene: association with recessive retinitis pigmentosa without hearing loss. Am J Hum Genet. 2000;66(6):1975-1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Estrada-Cuzcano A, Koenekoop RK, Coppieters F, et al. . IQCB1 mutations in patients with leber congenital amaurosis. Invest Ophthalmol Vis Sci. 2011;52(2):834-839. [DOI] [PubMed] [Google Scholar]
- 22.Xu M, Yang L, Wang F, et al. . Mutations in human IFT140 cause non-syndromic retinal degeneration. Hum Genet. 2015;134(10):1069-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Durlu YK, Köroğlu Ç, Tolun A. Novel recessive cone-rod dystrophy caused by POC1B mutation. JAMA Ophthalmol. 2014;132(10):1185-1191. [DOI] [PubMed] [Google Scholar]
- 24.Roosing S, Lamers IJ, de Vrieze E, et al. ; POC1B Study Group . Disruption of the basal body protein POC1B results in autosomal-recessive cone-rod dystrophy. Am J Hum Genet. 2014;95(2):131-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haer-Wigman L, Newman H, Leibu R, et al. . Non-syndromic retinitis pigmentosa due to mutations in the mucopolysaccharidosis type IIIC gene, heparan-alpha-glucosaminide N-acetyltransferase (HGSNAT). Hum Mol Genet. 2015;24(13):3742-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang X, Wang H, Sun V, et al. . Comprehensive molecular diagnosis of 179 Leber congenital amaurosis and juvenile retinitis pigmentosa patients by targeted next generation sequencing. J Med Genet. 2013;50(10):674-688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang F, Wang H, Tuan HF, et al. . Next generation sequencing-based molecular diagnosis of retinitis pigmentosa: identification of a novel genotype-phenotype correlation and clinical refinements. Hum Genet. 2014;133(3):331-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roosing S, van den Born LI, Sangermano R, et al. . Mutations in MFSD8, encoding a lysosomal membrane protein, are associated with nonsyndromic autosomal recessive macular dystrophy. Ophthalmology. 2015;122(1):170-179. [DOI] [PubMed] [Google Scholar]
- 29.McCulloch DL, Marmor MF, Brigell MG, et al. . ISCEV Standard for full-field clinical electroretinography (2015 update). Doc Ophthalmol. 2015;130(1):1-12. [DOI] [PubMed] [Google Scholar]
- 30.Hood DC, Bach M, Brigell M, et al. ; International Society For Clinical Electrophysiology of Vision . ISCEV standard for clinical multifocal electroretinography (mfERG) (2011 edition). Doc Ophthalmol. 2012;124(1):1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bach M, Brigell MG, Hawlina M, et al. . ISCEV standard for clinical pattern electroretinography (PERG): 2012 update. Doc Ophthalmol. 2013;126(1):1-7. [DOI] [PubMed] [Google Scholar]
- 32.Hull S, Arno G, Ku CA, et al. . Molecular and clinical findings in patients with Knobloch syndrome. JAMA Ophthalmol. 2016;134(7):753-762. [DOI] [PubMed] [Google Scholar]
- 33.Hull S, Owen N, Islam F, et al. . Nonsyndromic retinal dystrophy due to bi-allelic mutations in the ciliary transport gene IFT140. Invest Ophthalmol Vis Sci. 2016;57(3):1053-1062. [DOI] [PubMed] [Google Scholar]
- 34.Chiang JP, Lamey T, McLaren T, Thompson JA, Montgomery H, De Roach J. Progress and prospects of next-generation sequencing testing for inherited retinal dystrophy. Expert Rev Mol Diagn. 2015;15(10):1269-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller JN, Chan CH, Pearce DA. The role of nonsense-mediated decay in neuronal ceroid lipofuscinosis. Hum Mol Genet. 2013;22(13):2723-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.University College London NCL Resource: a gateway for Batten disease. http://www.ucl.ac.uk/ncl/. Accessed September 18, 2016.
- 37.Kohlschütter A, Schulz A. Towards understanding the neuronal ceroid lipofuscinoses. Brain Dev. 2009;31(7):499-502. [DOI] [PubMed] [Google Scholar]
- 38.Cotman SL, Staropoli JF. The juvenile Batten disease protein, CLN3, and its role in regulating anterograde and retrograde post-Golgi trafficking. Clin Lipidol. 2012;7(1):79-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garon ML, Rufiange M, Hamilton R, McCulloch DL, Lachapelle P. Asymmetrical growth of the photopic hill during the light adaptation effect. Doc Ophthalmol. 2010;121(3):177-187. [DOI] [PubMed] [Google Scholar]
- 40.Pinilla I, Lund RD, Sauvé Y. Contribution of rod and cone pathways to the dark-adapted electroretinogram (ERG) b-wave following retinal degeneration in RCS rats. Vision Res. 2004;44(21):2467-2474. [DOI] [PubMed] [Google Scholar]
- 41.Wiley LA, Burnight ER, Drack AV, et al. . Using patient-specific induced pluripotent stem cells and wild-type mice to develop a gene augmentation-based strategy to treat CLN3-associated retinal degeneration [published online July 11, 2016]. Hum Gene Ther. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ogura S, Yasukawa T, Kato A, et al. . Wide-field fundus autofluorescence imaging to evaluate retinal function in patients with retinitis pigmentosa. Am J Ophthalmol. 2014;158(5):1093-1098. [DOI] [PubMed] [Google Scholar]
- 43.Dulz S, Wagenfeld L, Nickel M, et al. . Novel morphological macular findings in juvenile CLN3 disease. Br J Ophthalmol. 2016;100(6):824-828. [DOI] [PubMed] [Google Scholar]
- 44.Robson AG, Egan CA, Luong VA, Bird AC, Holder GE, Fitzke FW. Comparison of fundus autofluorescence with photopic and scotopic fine-matrix mapping in patients with retinitis pigmentosa and normal visual acuity. Invest Ophthalmol Vis Sci. 2004;45(11):4119-4125. [DOI] [PubMed] [Google Scholar]
- 45.Robson AG, Tufail A, Fitzke F, et al. . Serial imaging and structure-function correlates of high-density rings of fundus autofluorescence in retinitis pigmentosa. Retina. 2011;31(8):1670-1679. [DOI] [PubMed] [Google Scholar]
- 46.Greenstein VC, Duncker T, Holopigian K, et al. . Structural and functional changes associated with normal and abnormal fundus autofluorescence in patients with retinitis pigmentosa. Retina. 2012;32(2):349-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Munroe PB, Mitchison HM, O’Rawe AM, et al. . Spectrum of mutations in the Batten disease gene, CLN3. Am J Hum Genet. 1997;61(2):310-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wisniewski KE, Zhong N, Kaczmarski W, et al. . Compound heterozygous genotype is associated with protracted juvenile neuronal ceroid lipofuscinosis. Ann Neurol. 1998;43(1):106-110. [DOI] [PubMed] [Google Scholar]
- 49.Lauronen L, Munroe PB, Järvelä I, et al. . Delayed classic and protracted phenotypes of compound heterozygous juvenile neuronal ceroid lipofuscinosis. Neurology. 1999;52(2):360-365. [DOI] [PubMed] [Google Scholar]
- 50.Sarpong A, Schottmann G, Rüther K, et al. . Protracted course of juvenile ceroid lipofuscinosis associated with a novel CLN3 mutation (p.Y199X). Clin Genet. 2009;76(1):38-45. [DOI] [PubMed] [Google Scholar]
- 51.Kelly JP, Weiss AH, Rowell G, Seigel GM. Autofluorescence and infrared retinal imaging in patients and obligate carriers with neuronal ceroid lipofuscinosis. Ophthalmic Genet. 2009;30(4):190-198. [DOI] [PubMed] [Google Scholar]
- 52.Holopainen JM, Brockman HL, Brown RE, Kinnunen PK. Interfacial interactions of ceramide with dimyristoylphosphatidylcholine: impact of the N-acyl chain. Biophys J. 2001;80(2):765-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim Y, Ramirez-Montealegre D, Pearce DA. A role in vacuolar arginine transport for yeast Btn1p and for human CLN3, the protein defective in Batten disease. Proc Natl Acad Sci U S A. 2003;100(26):15458-15462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Luiro K, Yliannala K, Ahtiainen L, et al. . Interconnections of CLN3, Hook1 and Rab proteins link Batten disease to defects in the endocytic pathway. Hum Mol Genet. 2004;13(23):3017-3027. [DOI] [PubMed] [Google Scholar]
- 55.Kyttälä A, Ihrke G, Vesa J, Schell MJ, Luzio JP. Two motifs target Batten disease protein CLN3 to lysosomes in transfected nonneuronal and neuronal cells. Mol Biol Cell. 2004;15(3):1313-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kyttälä A, Yliannala K, Schu P, Jalanko A, Luzio JP. AP-1 and AP-3 facilitate lysosomal targeting of Batten disease protein CLN3 via its dileucine motif. J Biol Chem. 2005;280(11):10277-10283. [DOI] [PubMed] [Google Scholar]
- 57.Metcalf DJ, Calvi AA, Seaman MNj, Mitchison HM, Cutler DF. Loss of the Batten disease gene CLN3 prevents exit from the TGN of the mannose 6-phosphate receptor. Traffic. 2008;9(11):1905-1914. [DOI] [PubMed] [Google Scholar]
- 58.Kama R, Kanneganti V, Ungermann C, Gerst JE. The yeast Batten disease orthologue Btn1 controls endosome-Golgi retrograde transport via SNARE assembly. J Cell Biol. 2011;195(2):203-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Herrmann P, Druckrey-Fiskaaen C, Kouznetsova E, et al. . Developmental impairments of select neurotransmitter systems in brains of Cln3(Deltaex7/8) knock-in mice, an animal model of juvenile neuronal ceroid lipofuscinosis. J Neurosci Res. 2008;86(8):1857-1870. [DOI] [PubMed] [Google Scholar]
- 60.Finn R, Kovács AD, Pearce DA. Altered sensitivity of cerebellar granule cells to glutamate receptor overactivation in the Cln3(Δex7/8)-knock-in mouse model of juvenile neuronal ceroid lipofuscinosis. Neurochem Int. 2011;58(6):648-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Katz ML, Gao CL, Prabhakaram M, Shibuya H, Liu PC, Johnson GS. Immunochemical localization of the Batten disease (CLN3) protein in retina. Invest Ophthalmol Vis Sci. 1997;38(11):2375-2386. [PubMed] [Google Scholar]
- 62.Uusi-Rauva K, Luiro K, Tanhuanpää K, et al. . Novel interactions of CLN3 protein link Batten disease to dysregulation of fodrin-Na+, K+ ATPase complex. Exp Cell Res. 2008;314(15):2895-2905. [DOI] [PubMed] [Google Scholar]
- 63.Wetzel RK, Arystarkhova E, Sweadner KJ. Cellular and subcellular specification of Na,K-ATPase alpha and beta isoforms in the postnatal development of mouse retina. J Neurosci. 1999;19(22):9878-9889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hofmann SL, Atashband A, Cho SK, Das AK, Gupta P, Lu JY. Neuronal ceroid lipofuscinoses caused by defects in soluble lysosomal enzymes (CLN1 and CLN2). Curr Mol Med. 2002;2(5):423-437. [DOI] [PubMed] [Google Scholar]
- 65.Narayan SB, Rakheja D, Tan L, Pastor JV, Bennett MJ. CLN3P, the Batten’s disease protein, is a novel palmitoyl-protein Delta-9 desaturase. Ann Neurol. 2006;60(5):570-577. [DOI] [PubMed] [Google Scholar]
- 66.Nosková L, Stránecký V, Hartmannová H, et al. . Mutations in DNAJC5, encoding cysteine-string protein alpha, cause autosomal-dominant adult-onset neuronal ceroid lipofuscinosis. Am J Hum Genet. 2011;89(2):241-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wavre-Shapton ST, Calvi AA, Turmaine M, et al. . Photoreceptor phagosome processing defects and disturbed autophagy in retinal pigment epithelium of Cln3Δex1-6 mice modelling juvenile neuronal ceroid lipofuscinosis (Batten disease). Hum Mol Genet. 2015;24(24):7060-7074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vidal-Donet JM, Cárcel-Trullols J, Casanova B, Aguado C, Knecht E. Alterations in ROS activity and lysosomal pH account for distinct patterns of macroautophagy in LINCL and JNCL fibroblasts. PLoS One. 2013;8(2):e55526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nugent T, Mole SE, Jones DT. The transmembrane topology of Batten disease protein CLN3 determined by consensus computational prediction constrained by experimental data. FEBS Lett. 2008;582(7):1019-1024. [DOI] [PubMed] [Google Scholar]
- 70.Kitzmüller C, Haines RL, Codlin S, Cutler DF, Mole SE. A function retained by the common mutant CLN3 protein is responsible for the late onset of juvenile neuronal ceroid lipofuscinosis. Hum Mol Genet. 2008;17(2):303-312. [DOI] [PubMed] [Google Scholar]
- 71.Chan CH, Mitchison HM, Pearce DA. Transcript and in silico analysis of CLN3 in juvenile neuronal ceroid lipofuscinosis and associated mouse models. Hum Mol Genet. 2008;17(21):3332-3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eTable. Functional prediction of CLN3 mutations associated with isolated retinal degeneration.
eFigure 1. Pedigrees and mutation segregation in 5 families for 6 patients.
eFigure 2. Supplementary full-field electroretinograms.
eFigure 3. Multifocal electroretinography of patients with late-onset retinal degeneration who retain residual full-field cone responses.
eFigure 4. Evaluation of the effect of intronic mutation c.461-3C>G on CLN3 transcript level and autofluorescent phenotype.
eFigure 5. Patients show electroretinography responses with reduced b-wave to a-wave ratio under scotopic conditions.