Key Points
Question
Does the cutaneous microbiome in hidradenitis suppurativa differ from that in healthy controls?
Findings
In this case-control study that included 30 patients with hidradenitis suppurativa and 24 healthy controls, next-generation sequencing analysis demonstrated a significantly different microbiome in patients with hidradenitis suppurativa (lesional and nonlesional) compared with that in healthy controls.
Meaning
Overall, the data suggest that a dysbiotic microbiome may have a role in the pathogenesis of hidradenitis suppurativa.
Abstract
Importance
Although the pathogenesis of hidradenitis suppurativa (HS) remains enigmatic, several factors point to potential involvement of the cutaneous microbiome. Insight into the cutaneous microbiome in HS using next-generation sequencing may provide novel data on the microbiological diversity of the skin.
Objective
To investigate the follicular skin microbiome in patients with HS and in healthy controls.
Design, Setting, and Participants
This case-control study obtained punch biopsy specimens from patients with HS (lesional and nonlesional) and healthy controls between October 1, 2014, and August 1, 2016. Data were analyzed from March to November 2016. Patients with HS were recruited from the Department of Dermatology, Zealand University Hospital, Roskilde, Denmark. Biopsy specimens were analyzed at the Department of Microbiology and Infection Control, Statens Serum Institut, Copenhagen, Denmark. None of the participants received any antibiotics (systemic or topical therapy) within 1 month before the study. In patients with HS, biopsy specimens were obtained from lesional skin (axilla or groin) and nonlesional skin. Only nodules containing at least 1 visible hair follicle were biopsied. Biopsy specimens from healthy controls were obtained from the axilla only.
Main Outcomes and Measures
The different microbiomes were investigated using next-generation sequencing targeting 16S and 18S ribosomal RNA.
Results
The skin microbiome was characterized in 30 patients with HS (mean [SD] age, 46.9 [14.0] years; 19 [63% female]) and 24 healthy controls (mean [SD] age, 32.2 [12.0] years; 13 [54% female]). The next-generation sequencing data provided a previously unreported (to our knowledge) characterization of the skin microbiome in HS. The study demonstrated that the microbiome in HS differs significantly from that in healthy controls in lesional and nonlesional skin. Overall, the following 5 microbiome types were identified: Corynebacterium species (type I), Acinetobacter and Moraxella species (type II), Staphylococcus epidermidis (type III), Porphyromonas and Peptoniphilus species (type IV), and Propionibacterium acnes (type V). In lesional skin, microbiome types consisted predominantly of type I or type IV. Microbiome type IV was not detected in healthy controls. Several taxa, including Propionibacterium, showed a significantly higher relative abundance in healthy controls vs HS skin, indicating that Propionibacterium may be part of the pathogenesis in HS.
Conclusions and Relevance
The study findings suggest a link between a dysbiotic cutaneous microbiome and HS.
This case-control study investigates the follicular skin microbiome in patients with hidradenitis suppurativa and healthy controls.
Introduction
Hidradenitis suppurativa (HS) is a chronic, inflammatory skin disease of the hair follicle defined by recurrent nodules, tunnels (sinus tracts), and scarring involving the intertriginous regions. The nodules may progressively expand to abscesses and subsequently rupture, causing suppuration and malodorous discharge. The prevalence of HS has been estimated to be among 1% to 4% of the European populations.
Although evidence implies that aberrant immune responses have a role involving the innate and adaptive immune systems, the clinical picture of HS lesions appears reminiscent of bacterial infection. However, the structure of the bacterial population, as well as the diversity at the strain level, is poorly investigated in patients with HS. Previous studies have been restricted to lesional skin, without inclusion of healthy control groups. Most previous bacteriological studies in HS have primarily relied on culture-based methods, and the results often suggested that commensal bacteria (eg, coagulase-negative staphylococci) have a role in HS.
The emergence of next-generation sequencing (NGS) has allowed a more accurate and less biased characterization of microbiomes compared with culture-based techniques. Therefore, it appears attractive to investigate the microbiome of HS using NGS, thereby yielding a wider spectrum of species. We designed a case-control study investigating the skin microbiome using NGS targeting 16S and 18S ribosomal RNA (rRNA). Regions within the 6SrRNA gene give information on species-specific signature sequences, thereby providing valuable data for bacteria (16S) and for the identification of other organisms (18S) (eg, fungi). Insight into the cutaneous microbiome in HS using NGS may provide novel data on the microbiological diversity of the skin.
Methods
Ethical Statement and Study Design
This study was approved by the Ethics Committee of Region Zealand (project SJ-420) and the data protection agency (REG-105-2014) in Denmark. The study was conducted at the Department of Dermatology, Zealand University Hospital, Roskilde, Denmark, from October 1, 2014, to August 1, 2016. Data were analyzed from March to November 2016. Written informed consent was obtained from all study participants. An exploratory case-control study was designed to compare individuals with the diagnosis of HS (n = 30) with healthy controls (n = 24).
Study Participants
Participants with HS were randomly selected by consecutive recruitment of eligible patients from the Department of Dermatology, Zealand University Hospital. Healthy controls were recruited from the University of Copenhagen and from the health care staff at Zealand University Hospital.
Inclusion Criteria
All cases had a verified diagnosis of HS (International Statistical Classification of Diseases, 10th Revision, code L73.2) at the Department of Dermatology, Zealand University Hospital. An experienced dermatologist (D.M.S.) diagnosed all included patients. To verify the diagnosis of HS, histological biopsy specimens from lesional skin were obtained from all included patients. Furthermore, all patients were 18 years or older, were not pregnant, and experienced a flare of at least 1 inflamed nodule located in the axilla or in the groin.
Exclusion Criteria
Exclusion criteria were treatment with any antibiotics (systemic or topical therapy) within 1 month before the study (patients and healthy controls). Healthy controls were examined and excluded if any lesions compatible with HS were found. Healthy controls were also excluded if they had any other skin manifestations that would affect sampling or the microbial community at the time of sampling (a discussion of HS severity is available in the eAppendix in the Supplement).
Skin Biopsies
Before injection of anesthetics, the skin was cleansed with ethanol (70% isopropyl alcohol) swabs. The HS lesional and nonlesional skin was anesthetized using a solution of lidocaine hydrochloride (20 mg/mL), and adrenalin (5 µg/mL), with no preservatives. In patients with HS, one 4-mm punch biopsy specimen was obtained from lesional skin (axilla or groin) and nonlesional skin. Biopsy specimens from HS nonlesional skin were obtained approximately 5 cm from the inflamed nodule and were clinically unaffected.
Only nodules containing at least 1 visible hair follicle were biopsied in all participants. Biopsy specimens from healthy controls were obtained from the axilla only.
Microbiome Sequencing and Analysis
Biopsy specimens were analyzed at the Department of Microbiology and Infection Control, Statens Serum Institut, Copenhagen, Denmark. DNA was extracted using a kit (QIAamp DNA Mini Kit; Qiagen) according to the manufacturer’s instructions for tissues. For each batch of DNA extraction, a “negative” control was included containing buffers but no sample material for downstream analysis. DNA was amplified using a 2-step polymerase chain reaction using custom 341F/806R primers targeting the V3-V4 16S regions, as well as 3 primer sets targeting the hypervariable regions V3-V4 of the 18SrDNA gene, and amplicons were sequenced on a desktop sequencer (MiSeq; Illumina, Inc) using the v2 reagent kit. For details concerning primer design and library preparation, see the eAppendix in the Supplement. Sequence data are available at the European Nucleotide Archive (accession number PRJEB15266).
Statistical Analysis
All data analysis was conducted using a statistical software package (R, version 3.2.3; The R Project for Statistical Computing). Microbiome data were handled using the add-on package “phyloseq” version 1.16.2 and visualized with “ggplot2” version 2.1.0. Microbiome types were defined using vegdist (from the add-on package “vegan” version 2.3-2) and hierarchical clustering with default settings, and the number of optimal groups was decided by manual inspection.
Differences between groups were assessed with bar plots and principal coordinates analysis plots and tested with Kruskal-Wallis test or permutation test. Differential abundances (DAs) were tested using linear models, adjusted for anatomical location (axilla vs groin). Statistical analyses were performed for associations between the 5 microbiome types (Corynebacterium species [type I], Acinetobacter and Moraxella species [type II], Staphylococcus epidermidis [type III], Porphyromonas and Peptoniphilus species [type IV], and Propionibacterium acnes [type V]) in the 3 groups (HS lesional skin, HS nonlesional skin, and healthy controls) and the variables sex, age, body mass index (BMI), smoking status, Sartorius score, and Hurley stage using Fisher exact test for categorical variables and Kruskal-Wallis test for continuous variables. A full statistical description is available in the eAppendix in the Supplement.
Results
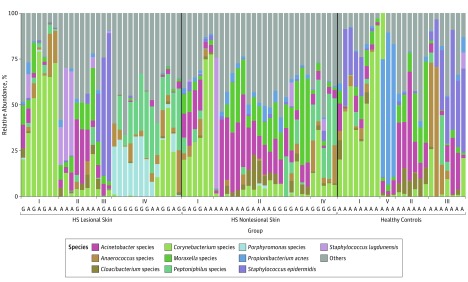
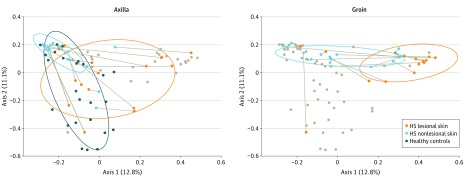
The cutaneous microbiome was characterized in 30 patients with HS and in 24 healthy controls. In total, 83 punch biopsy specimens (30 HS lesional skin, 29 HS nonlesional skin, and 24 healthy controls) were obtained from patients with HS and healthy controls. Background factors and characteristics listed in the Table show that most of the patients with HS were predominantly female, younger, and smokers. Overall, our data demonstrated that the microbiome in inflamed HS nodules, nonlesional skin, and samples from healthy controls differed significantly from each other (Figure 1, Figure 2, and the eTable in the Supplement). The bar plot in Figure 1 shows differences in the bacterial community, with the 10 most abundant species or genera across the 3 groups. Several different unique types (I-V) of the skin microbiome were defined using hierarchical clustering of Bray-Curtis distances (Figure 1), a common dissimilarity metric used in ecology. Microbiome types in lesional skin consisted predominantly of Corynebacterium species (type I) and Porphyromonas and Peptoniphilus species (type IV). In contrast, Acinetobacter and Moraxella species (type II) dominated nonlesional skin. In healthy controls, the various microbiome types (I, II, III, and V) were more equally distributed. Microbiome types III and V were dominated by S epidermidis and P acnes, respectively. Microbiome type IV was not detected in healthy controls, and microbiome type V was not found in HS lesional or nonlesional skin. The principal coordinates analysis plots (Figure 2 and eFigure 1 in the Supplement) show the significant differences between the 3 groups (overall F = 3.67 [between-group variance vs within-group variance, values above 1 indicate separation], P < .001; lesional vs non-lesional, F = 3.11, P < .001; lesional vs healthy controls, F = 4.21, P < .001; nonlesional vs healthy controls F = 3.65, P < .001, by Adonis PERMANOVA test) with and without stratification for anatomical location. There was no significant association between the number of species and the duration of lesions or the diameter of lesions.
Table. Background Characteristics of Study Participants.
| Characteristic | Patients With HS | Healthy Controls |
|---|---|---|
| Total analyzed | 30 | 24 |
| Female, No. (%) | 19 (63) | 13 (54) |
| Age, mean (SD), ya | 46.9 (14.0) | 32.2 (12.0) |
| White race/ethnicity, No. (%) | 30 (100) | 23 (96) |
| BMI, mean (SD)a | 30.5 (7.7) | 26.5 (2.6) |
| Smoking status, No. (%)b | 20 (67) | 7 (29) |
| Sartorius score, mean (SD) | 24 (20) | NA |
| HS severity, No. (%) | ||
| Hurley stage 1 | 0 | NA |
| Hurley stage 2 | 26 (87) | NA |
| Hurley stage 3 | 4 (13) | NA |
| Duration of lesions, median (IQR), d | 21 (14-60) | NA |
| Location of lesions | Groin (n = 15), axilla (n = 15) | Axilla |
| Diameter of lesions, mean (SD), cm | 2.76 (1.38) | NA |
Abbreviations: BMI, body mass index (calculated as weight in kilograms divided by height in meters squared); HS, hidradenitis suppurativa; IQR, interquartile range; NA, not applicable.
P < .05 by Wilcoxon rank sum test.
P < .05 by χ2 test.
Figure 1. Distribution of the Top 10 Most Abundant Species Found in Patients With Hidradenitis Suppurativa (HS) and in Healthy Controls.
Shown is the distribution of the 10 most abundant species found in HS lesional skin, HS nonlesional skin, and healthy controls. Overall, the following 5 microbiome types were identified: Corynebacterium species (type I), Acinetobacter and Moraxella species (type II), Staphylococcus epidermidis (type III), Porphyromonas and Peptoniphilus species (type IV), and Propionibacterium acnes (type V). A and G indicate anatomical location of the samples (axilla or groin).
Figure 2. Bray-Curtis Principal Coordinates Analysis Plot Showing Differences Between the 3 Groups.
Shown are differences between the 3 groups (HS lesional skin, HS nonlesional skin, and healthy controls) in axillary and groin samples. Each lesional sample is connected with its corresponding nonlesional sample. Gray dots indicate samples from the opposite anatomical location (axilla or groin). Ellipses indicate the 75% prediction areas of samples from each group. HS indicates hidradenitis suppurativa.
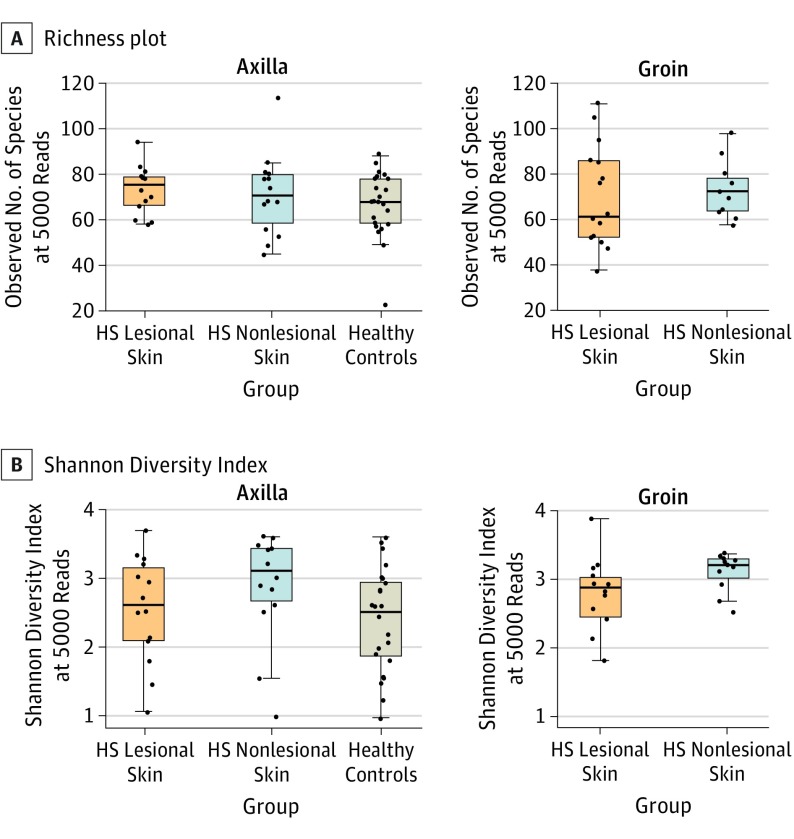
In the 3 groups, no difference in richness (number of species per sample) was found (HS lesional median [IQR] 72 [59-82], HS nonlesional 72 [63-80], healthy controls 68 [59-78], overall P = .66, HS lesional skin vs HS nonlesional skin P = .97, HS lesional skin vs healthy controls P = .46, and HS nonlesional skin vs healthy controls P = .41) (Figure 3A). However, as shown in Figure 3B, an increased Shannon Diversity Index (a combined measure of evenness and number of species) due to increased taxonomic evenness (evenness between species) was found in nonlesional skin (median [IQR] 3.21 [2.84-3.39]) compared with lesional skin (2.80 [2.14-3.09], P = .02) and healthy controls (2.52 [1.88-2.95], P = .003) (overall P = .005), while lesional skin and healthy controls were not significantly different (P = .22). No interactions were found between the sample groups and anatomical location (linear models with vs without interaction terms; Richness P = .54, Shannon Index P = .91).
Figure 3. Richness Plot and Shannon Diversity Index.
A, Richness plot stratified by anatomical location shows no difference between the number of species per sample in the 3 groups (HS lesional skin median [IQR] 72 [59-82], HS nonlesional skin 72 [63-80], and healthy controls 68 [59-78], P = .66) after rarefaction to 5000 reads. No difference was found between the 2 anatomical locations (model comparison with vs without interaction term between group and location, P = .54). B, Shannon Diversity Index stratified by anatomical location shows the diversity of the species in each group. The index reveals an increased diversity in HS nonlesional skin (median [IQR] 3.21 [2.84-3.39]) compared with HS lesional skin and healthy controls (2.80 [2.14-3.09] and 2.52 [1.88-2.95], overall P = .005). No difference was found between the 2 anatomical locations (model comparison with vs without interaction terms between group and location, P = .91). HS indicates hidradenitis suppurativa.
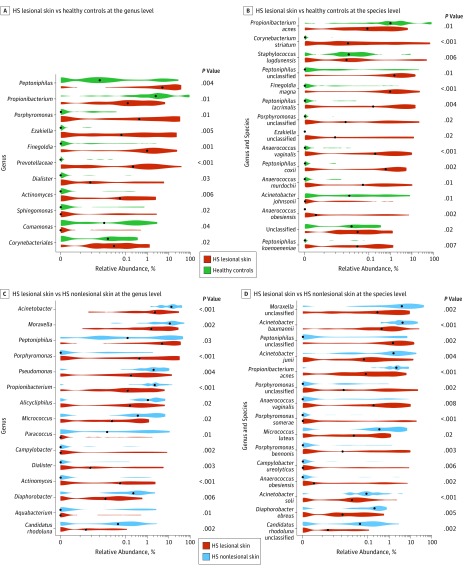
The DA analyses at the genus and species levels yielded various differences between groups (Figure 4). At the genus level, the DA analysis showed a significantly increased relative abundance of Porphyromonas and Peptoniphilus species (abundant in microbiome type IV) in lesional skin samples vs healthy control samples (Figure 4A). Furthermore, Propionibacterium species showed a significantly higher relative abundance in healthy controls vs lesional skin (Figure 4A). At the species level, a significantly higher relative abundance of P acnes and Corynebacterium striatum was found in the healthy control group compared with that in HS lesional skin (Figure 4B). This finding is also in congruence with the bar plot (Figure 1) showing microbiome type V (P acnes) as unique for healthy controls.
Figure 4. Differential Abundance Analyses.
A, Differential abundance at genus level between hidradenitis suppurativa (HS) lesional skin and healthy controls. Note significantly higher relative abundance of Propionibacterium in healthy controls and significantly higher relative abundance of Porphyromonas and Peptoniphilus. B, Differential abundance at species level between HS lesional skin and healthy controls. Note higher relative abundance of Propionibacterium acnes in healthy controls than in lesional samples. C, Differential abundance at genus level between HS lesional skin and HS nonlesional skin. Note significantly higher relative abundance of Porphyromonas and Peptoniphilus in lesional skin than in nonlesional skin, Propionibacterium is more abundant in nonlesional skin. D, Differential abundance at species level between HS lesional skin and HS nonlesional skin. Note significantly reduced relative abundance of Propionibacterium acnes in HS lesional skin; higher relative abundances of Porphyromonas and Peptoniphilus are found in lesional skin. P values listed are adjusted for anatomical location. Black dots indicate the medians of each distribution.
In lesional skin vs nonlesional skin, the relative abundance of Porphyromonas and Peptoniphilus species remained significantly higher in lesional skin at the genus and species levels (Figure 4C and D). Conversely, a significantly reduced relative abundance of Propionibacterium species and P acnes was found in lesional skin vs nonlesional skin (Figure 4C and D). The DA analysis of nonlesional skin vs healthy controls did not yield any significant associations after controlling for the false discovery rate.
In lesional skin, a significant association was found between microbiome types and location (Fischer exact test, P = .04), with biopsy specimens obtained from the groin showing a higher frequency of microbiome type IV (groin 67% [n = 10] vs axilla 20% [n = 3], see eTable in the Supplement). In nonlesional skin, a significant association was also found between microbiome types and location (Fischer exact test, P = .04), with microbiome type II associated with biopsy specimens obtained from the axilla (80% [n = 12] vs groin 43% [n = 6]); again, we found microbiome type IV to be associated with biopsy specimens obtained from the groin (36% [n = 5] vs axilla 0% [n = 0]). In nonlesional skin, but not in lesional skin, sex had a significant association with microbiome type (Fischer exact test, P = .04): female patients were more likely to have microbiome type II (72% [n = 13] vs males 45% [n = 5]). There was no difference in the distribution of axillary and groin biopsy specimens between the sexes (females 42% [n = 8] axilla samples vs males 64% [n = 7], P = .45). In patients with HS, no significant association was found between microbiome types and some variables, including age, BMI, smoking status, Sartorius score, and diameter of lesions. However, when correlating medication use (resorcinol monoacetate or azelaic acid) with the various microbiome types, medication was found to be associated with type I (treated 50% [n = 5] vs nontreated 5% [n = 1], P = .01) in nonlesional samples. In healthy controls, no significant association was found between sex, age, BMI, smoking status, and microbiome types.
Examining the correlation between beta diversity and these factors in lesional samples, we found a difference between biopsy locations (F = 2.05, P = .01), but not due to BMI (F = 0.49, P = .99), topical treatment (F = 1.07, P = .31), or smoking status (F = 0.85, P = .64) (eFigure 2A, eFigure 2B, and eFigure 2C in the Supplement). Stratified analyses showed that differences observed between groups were not confounded but remained significant despite stratification for these covariates (BMI<30 F = 2.05, P < .001; axilla only F = 2.20, P < .001; no treatment F = 3.25, P < .001) (eFigure 3A, eFigure 3B, and eFigure 3C in the Supplement).
The 18S analysis was also performed in Malassezia species. The analysis yielded no significant differences between groups.
Discussion
Overall, our NGS approach provided a previously unreported (to our knowledge) characterization of the skin microbiome in HS. The data demonstrated that the microbiome in HS differs significantly from that in healthy controls in HS lesional and nonlesional skin (Figures 1 and 2). This difference remained significant when stratifying for anatomical location of the samples (Figure 2). In addition, the DA analyses (Figure 4) showed significant differences at the genus and species levels between groups. Surprisingly, there were no differences in richness, the overall number of species found in each group (Figure 3A). However, in nonlesional skin, a significantly increased Shannon Diversity Index, which we interpret as increased taxonomic evenness, was found in axillary and groin samples (Figure 3B), indicating a possible imbalance (dysbiosis) of the microbial community in clinically normal–appearing HS skin.
The presence of anaerobic bacteria, such as Porphyromonas and Peptoniphilus species (type IV), has previously been reported in HS. Both bacteria belong to the spectrum of commensal bacteria on mucocutaneous surfaces. Guet-Revillet et al recently investigated the microbiological profiles of 102 HS lesions using matrix-assisted laser desorption–time-of-flight mass spectrometry. In mature lesions, the study identified these types of bacteria (Porphyromonas and Peptoniphilus species) in 10 and 15 isolates, respectively. Both bacteria have previously been found in the vaginal flora. This result is in congruence with our findings of an association of microbiome type IV and the groin, where secondary colonization from the vaginal flora or intimate contact with other partners may hypothetically occur. Alternatively, in particular, Porphyromonas species may have reached the groin indirectly from the oral cavity. Nevertheless, both bacteria have been described in chronic wounds and skin abscesses and may contribute to the chronicity and prolonged inflammation in HS lesions. Although we identified these species in inflamed HS nodules only, and not in chronic suppurating lesions, these species may have a role in the pathogenesis of HS at a much earlier event than previously considered. However, considering the intrapersonal variations of the skin microbiome between different skin sites, a comparison with the healthy controls may be somewhat biased because we did not obtain biopsy specimens from the groin among healthy controls. This absence may potentially have contributed to the lack of microbiome type IV in healthy controls. However, we found that the largest source of variation, even within individuals, was indeed the sample groups, as shown in Figure 2. Therefore, the potential dysbiosis in HS skin seems to outweigh any effects of the normal biogeographic diversity in the skin.
Although microbiome type IV was primarily associated with HS groin samples, Porphyromonas and Peptoniphilus species are indeed not considered part of the normal cutaneous microbiota in the human groin. Most important, the normal human axillary and groin microbiota are predominantly composed of Corynebacterium species, Propionibacterium species, and staphylococci species.
Although P acnes has a well-described pathogenic role in acne vulgaris, it also serves as an abundant skin commensal that offers bactericidal properties against several other pathogens. In the perspective of the potential beneficial role of P acnes, the significant reduced relative abundance of P acnes in HS lesional skin compared with healthy controls may give rise to speculations on its clinical relevance. The potential underlying mechanism of this observation may reflect the pathogenic role of sebaceous glands in HS. Although we did not find a reduced presence of Malassezia species in patients with HS, using stereology, Kamp et al showed that sebaceous gland number and volume in patients with HS were significantly reduced compared with healthy persons. Furthermore, sebum excretion has also previously been shown to be reduced in HS. Most important, sebaceous glands are anoxic, and the secretion of sebum supports the proliferation of facultative anaerobic bacteria, such as P acnes. Therefore, the association between the reduction in number or volume in sebaceous glands and the subsequent alterations in the normal skin microbiota (eg, reduced presence of P acnes) may be part of the pathogenesis of HS.
In nonlesional skin from patients with HS, we found a significantly increased diversity of the microbiota compared with HS lesional skin and healthy controls, while the overall number of species was the same, indicating that HS nonlesional skin is characterized by a higher taxonomic evenness of several species. Although this result may reflect only the normal biological homogeneous distribution of the microbiota in inverse areas, this finding lends credence to a hypothesis of an imbalanced skin microbiome preceding the development of HS lesions. This hypothesis may also be further supported by a 2012 study demonstrating alterations in leukocyte subsets (low-grade leukocytic infiltration) and histomorphology (follicular plugging) in normal-appearing HS perilesional skin. However, whether this subclinical inflammation and histomorphological changes occur before the development of the potential dysbiosis can only be speculated.
Our group recently demonstrated a significantly reduced presence of microbiota in preclinical HS skin compared with that in healthy controls using peptide nucleic acid–fluorescence in situ hybridization (PNA-FISH). The healthy controls showed a significantly higher presence of bacterial aggregates (biofilms) of a cocci morphology. All the HS biopsy specimens in the PNA-FISH study appeared as clinically normal–appearing skin with visible hair follicles. Most important, however, all the patients with HS reported a history of intermittent disease activity at the investigated site. With regard to the present NGS study, we did not obtain information on previous disease activity in the nonlesional area, and not all nonlesional biopsy specimens contained visible hair follicles. Therefore, the nonlesional skin in this NGS study and the preclinical skin in the PNA-FISH study may be not biologically comparable. Furthermore, NGS yields relative abundances, not absolute abundances, meaning that the data cannot be compared directly with a quantitative measure of biomass, such as PNA-FISH. However, HS is considered a chronic and systemic disease, with an unpredictable location of the lesions within the predilection regions. Therefore, the presence of bacteria consistent with a cocci morphology found in healthy controls (microbiome type III), but not in HS nonlesional skin, seems in agreement with the PNA-FISH findings.
Strengths and Limitations
The present study has several strengths. To our knowledge, no previous case-control studies have been conducted on the microbiological profiles of HS using NGS. Moreover, because HS is linked to pathogenic events of intertriginous hair follicles and due to the fact that bacteria are present in subepidermal compartments, we obtained biopsy specimens containing visible hair follicles only. In addition, biopsy samples were taken only from inflamed nodules, which potentially provides information on the early pathogenesis of HS. Last, no included participants received any antibiotics (systemic or local) within 1 month before the time of investigation.
However, potential limitations may also have influenced our results. Overall, microbiomes may vary by community, potentially limiting the generalizability. The few participants may have affected the statistical analysis and limited the generalizability to the larger HS population. We did not investigate chronic suppurating lesions. Furthermore, local keratolytic agents may have affected the follicular skin microbiome in 9 patients (eTable in the Supplement). Although we used ethanol swabs before performing biopsies, we did not obtain information on personal hygiene, which may affect the microbial community. In addition, we did not investigate the microbiome over time; therefore, our data represent points in a timeline, which may display fluctuations.
Conclusions
The observation that the microbiome in HS lesional and nonlesional skin differs significantly from that in healthy controls supports the hypothesis of a link between a dysbiotic cutaneous microbiome and HS. More studies are warranted to definitively assign a causative role for the cutaneous microbiome in HS.
eAppendix. Supplemental Appendix
eTable. Distribution of Microbiome Types and Association With Location and Medication
eFigure 1. PCoA Plot Illustrating Differences Between the 3 Groups
eFigure 2A. Bray-Curtis PCoA, HS Lesional Skin, Axilla vs Groin
eFigure 2B. Bray-Curtis PCoA, HS Lesional Skin, High vs Low BMI
eFigure 2C. Bray-Curtis PCoA, HS Lesional Skin, Topical Treatment vs No Treatment
eFigure 3A. Bray-Curtis PCoA, HS Lesional Skin, Axilla Samples Only
eFigure 3B. Bray-Curtis PCoA, HS Lesional Skin, Low BMI Only
eFigure 3C. Bray-Curtis PCoA, HS Lesional Skin, No Topical Treatment
References
- 1.Zouboulis CC, Del Marmol V, Mrowietz U, Prens EP, Tzellos T, Jemec GB. Hidradenitis suppurativa/acne inversa: criteria for diagnosis, severity assessment, classification and disease evaluation. Dermatology. 2015;231(2):184-190. [DOI] [PubMed] [Google Scholar]
- 2.Jemec GB. Clinical practice: hidradenitis suppurativa. N Engl J Med. 2012;366(2):158-164. [DOI] [PubMed] [Google Scholar]
- 3.Jemec GB, Heidenheim M, Nielsen NH. The prevalence of hidradenitis suppurativa and its potential precursor lesions. J Am Acad Dermatol. 1996;35(2, pt 1):191-194. [DOI] [PubMed] [Google Scholar]
- 4.Vinding GR, Miller IM, Zarchi K, Ibler KS, Ellervik C, Jemec GB. The prevalence of inverse recurrent suppuration: a population-based study of possible hidradenitis suppurativa. Br J Dermatol. 2014;170(4):884-889. [DOI] [PubMed] [Google Scholar]
- 5.Jemec GB, Kimball AB. Hidradenitis suppurativa: epidemiology and scope of the problem. J Am Acad Dermatol. 2015;73(5)(suppl 1):S4-S7. [DOI] [PubMed] [Google Scholar]
- 6.van der Zee HH, de Ruiter L, van den Broecke DG, Dik WA, Laman JD, Prens EP. Elevated levels of tumour necrosis factor (TNF)-α, interleukin (IL)-1β and IL-10 in hidradenitis suppurativa skin: a rationale for targeting TNF-α and IL-1β. Br J Dermatol. 2011;164(6):1292-1298. [DOI] [PubMed] [Google Scholar]
- 7.Giamarellos-Bourboulis EJ, Antonopoulou A, Petropoulou C, et al. Altered innate and adaptive immune responses in patients with hidradenitis suppurativa. Br J Dermatol. 2007;156(1):51-56. [DOI] [PubMed] [Google Scholar]
- 8.Ring HC, Riis Mikkelsen P, Miller IM, et al. The bacteriology of hidradenitis suppurativa: a systematic review. Exp Dermatol. 2015;24(10):727-731. [DOI] [PubMed] [Google Scholar]
- 9.Sartorius K, Killasli H, Oprica C, Sullivan A, Lapins J. Bacteriology of hidradenitis suppurativa exacerbations and deep tissue cultures obtained during carbon dioxide laser treatment. Br J Dermatol. 2012;166(4):879-883. [DOI] [PubMed] [Google Scholar]
- 10.Lapins J, Jarstrand C, Emtestam L. Coagulase-negative staphylococci are the most common bacteria found in cultures from the deep portions of hidradenitis suppurativa lesions, as obtained by carbon dioxide laser surgery. Br J Dermatol. 1999;140(1):90-95. [DOI] [PubMed] [Google Scholar]
- 11.Brook I, Frazier EH. Aerobic and anaerobic microbiology of axillary hidradenitis suppurativa. J Med Microbiol. 1999;48(1):103-105. [DOI] [PubMed] [Google Scholar]
- 12.Jemec GB, Faber M, Gutschik E, Wendelboe P. The bacteriology of hidradenitis suppurativa. Dermatology. 1996;193(3):203-206. [DOI] [PubMed] [Google Scholar]
- 13.Matusiak Ł, Bieniek A, Szepietowski JC. Bacteriology of hidradenitis suppurativa: which antibiotics are the treatment of choice? Acta Derm Venereol. 2014;94(6):699-702. [DOI] [PubMed] [Google Scholar]
- 14.Nikolakis G, Join-Lambert O, Karagiannidis I, Guet-Revillet H, Zouboulis CC, Nassif A. Bacteriology of hidradenitis suppurativa/acne inversa: a review. J Am Acad Dermatol. 2015;73(5)(suppl 1):S12-S18. [DOI] [PubMed] [Google Scholar]
- 15.Buchan BW, Ledeboer NA. Emerging technologies for the clinical microbiology laboratory. Clin Microbiol Rev. 2014;27(4):783-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardie JM. Oral microbiology: current concepts in the microbiology of dental caries and periodontal disease. Br Dent J. 1992;172(7):271-278. [DOI] [PubMed] [Google Scholar]
- 17.Murdoch DA. Gram-positive anaerobic cocci. Clin Microbiol Rev. 1998;11(1):81-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guet-Revillet H, Coignard-Biehler H, Jais JP, et al. Bacterial pathogens associated with hidradenitis suppurativa, France. Emerg Infect Dis. 2014;20(12):1990-1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Albert AY, Chaban B, Wagner EC, et al. ; VOGUE Research Group . A study of the vaginal microbiome in healthy Canadian women utilizing cpn60-based molecular profiling reveals distinct Gardnerella subgroup community state types. PLoS One. 2015;10(8):e0135620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hill JE, Goh SH, Money DM, et al. Characterization of vaginal microflora of healthy, nonpregnant women by chaperonin-60 sequence-based methods. Am J Obstet Gynecol. 2005;193(3, pt 1):682-692. [DOI] [PubMed] [Google Scholar]
- 21.Mysak J, Podzimek S, Sommerova P, et al. Porphyromonas gingivalis: major periodontopathic pathogen overview. J Immunol Res. 2014;2014:476068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murphy EC, Frick IM. Gram-positive anaerobic cocci: commensals and opportunistic pathogens. FEMS Microbiol Rev. 2013;37(4):520-553. [DOI] [PubMed] [Google Scholar]
- 23.Brook I. Prevotella and Porphyromonas infections in children. J Med Microbiol. 1995;42(5):340-347. [DOI] [PubMed] [Google Scholar]
- 24.Callewaert C, Kerckhof FM, Granitsiotis MS, Van Gele M, Van de Wiele T, Boon N. Characterization of Staphylococcus and Corynebacterium clusters in the human axillary region. PLoS One. 2013;8(8):e70538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grice EA, Segre JA. The skin microbiome [published correction appears in Nat Rev Microbiol. 201;9(8):626]. Nat Rev Microbiol. 2011;9(4):244-253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grice EA, Kong HH, Conlan S, et al. ; NISC Comparative Sequencing Program . Topographical and temporal diversity of the human skin microbiome. Science. 2009;324(5931):1190-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fitz-Gibbon S, Tomida S, Chiu BH, et al. Propionibacterium acnes strain populations in the human skin microbiome associated with acne. J Invest Dermatol. 2013;133(9):2152-2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanford JA, Gallo RL. Functions of the skin microbiota in health and disease. Semin Immunol. 2013;25(5):370-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kamp S, Fiehn AM, Stenderup K, et al. Hidradenitis suppurativa: a disease of the absent sebaceous gland? sebaceous gland number and volume are significantly reduced in uninvolved hair follicles from patients with hidradenitis suppurativa. Br J Dermatol. 2011;164(5):1017-1022. [DOI] [PubMed] [Google Scholar]
- 30.Jemec GB, Gniadecka M. Sebum excretion in hidradenitis suppurativa. Dermatology. 1997;194(4):325-328. [DOI] [PubMed] [Google Scholar]
- 31.van der Zee HH, de Ruiter L, Boer J, et al. Alterations in leucocyte subsets and histomorphology in normal-appearing perilesional skin and early and chronic hidradenitis suppurativa lesions. Br J Dermatol. 2012;166(1):98-106. [DOI] [PubMed] [Google Scholar]
- 32.Ring HC, Bay L, Kallenbach K, et al. Normal skin microbiota is altered in pre-clinical hidradenitis suppurativa. Acta Derm Venereol. 2017;97(2):208-213. [DOI] [PubMed] [Google Scholar]
- 33.Jemec GB, Hansen U. Histology of hidradenitis suppurativa. J Am Acad Dermatol. 1996;34(6):994-999. [DOI] [PubMed] [Google Scholar]
- 34.von Laffert M, Helmbold P, Wohlrab J, Fiedler E, Stadie V, Marsch WC. Hidradenitis suppurativa (acne inversa): early inflammatory events at terminal follicles and at interfollicular epidermis. Exp Dermatol. 2010;19(6):533-537. [DOI] [PubMed] [Google Scholar]
- 35.Nakatsuji T, Chiang HI, Jiang SB, Nagarajan H, Zengler K, Gallo RL. The microbiome extends to subepidermal compartments of normal skin. Nat Commun. 2013;4:1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Callewaert C, Hutapea P, Van de Wiele T, Boon N. Deodorants and antiperspirants affect the axillary bacterial community. Arch Dermatol Res. 2014;306(8):701-710. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eAppendix. Supplemental Appendix
eTable. Distribution of Microbiome Types and Association With Location and Medication
eFigure 1. PCoA Plot Illustrating Differences Between the 3 Groups
eFigure 2A. Bray-Curtis PCoA, HS Lesional Skin, Axilla vs Groin
eFigure 2B. Bray-Curtis PCoA, HS Lesional Skin, High vs Low BMI
eFigure 2C. Bray-Curtis PCoA, HS Lesional Skin, Topical Treatment vs No Treatment
eFigure 3A. Bray-Curtis PCoA, HS Lesional Skin, Axilla Samples Only
eFigure 3B. Bray-Curtis PCoA, HS Lesional Skin, Low BMI Only
eFigure 3C. Bray-Curtis PCoA, HS Lesional Skin, No Topical Treatment