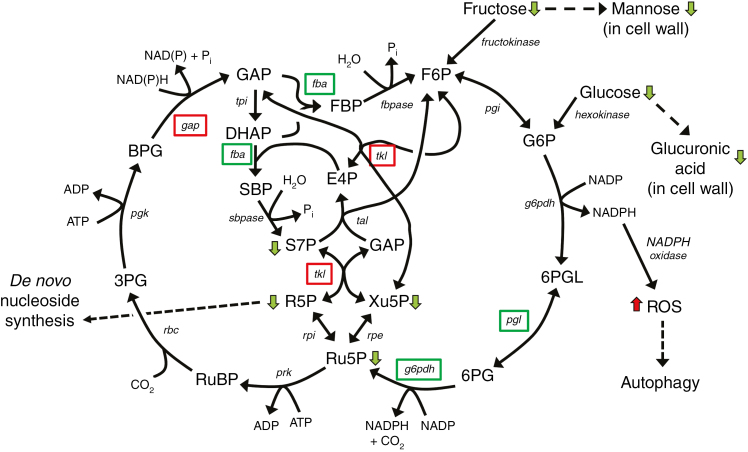
Fig. 8.
Model of the effect of the rns2-2 mutation on arabidopsis metabolism. Integration of transcriptome and metabolome data suggests that carbon flux is shuttled through the PPP. Since rns2-2 plants are deficient in rRNA turnover and nucleotide salvage, the increased carbon flux through the PPP pathway may be needed to generate ribose-5-phosphate for de novo synthesis of nucleotides to maintain cellular homeostasis. Changes in metabolism also result in increased NADPH levels. This increase causes accumulation of reactive oxygen species (ROS) that in turn signal the activation of the autophagy machinery. Green boxes indicate enzymes encoded by a gene downregulated in rns2-2, and red boxes upregulated. Green and red arrows indicate metabolites with lower and higher levels in rns2-2, respectively. F6P, fructose-6-phosphate; G6P, glucose-6-phosphate; 6PGL, 6-phosphogluconolactone; 6PG, 6-phosphogluconate; Ru5P, ribulose-5-phosphate; Xu5P, xylulose-5-phosphate; GAP, glyceraldehyde-3-phosphate; E4P, erythrose-4-phosphate; R5P, ribose-5-phosphate; S7P, sedoheptulose-7-phosphate; RuBP, ribulose-1,5-bisphosphate; 3PG, 3-phosphoglycerate; BPG, 1,3-bisphosphoglycerate; DHAP, dihydroxyacetone-phosphate; SBP, sedoheptulose-1,7-bisphosphate; FBP, fructose-1,6-bisphosphate; pgi, glucose phosphate isomerase; g6pdh, glucose-6-phosphate dehydrogenase; pgl, 6-phosphogluconolactonase; rpe, ribulose-5-phosphate epimerase; tal, transaldolase; tkl, transketolase; fbpase, fructose-2,6-bisphosphatase; fba, fructose-bisphosphate aldolase; rpi, ribose-5-phosphate isomerase; sbpase, sedoheptulose-1,7-bisphosphatase; tpi, triosephosphate isomerase; gap, glyceraldehyde-3-phosphate dehydrogenase; pgk, phosphoglycerate kinase; rbc, ribulose-1,5-bisphosphate carboxylase/oxygenase; prk, phosphoribulokinase.