Fig. 2.
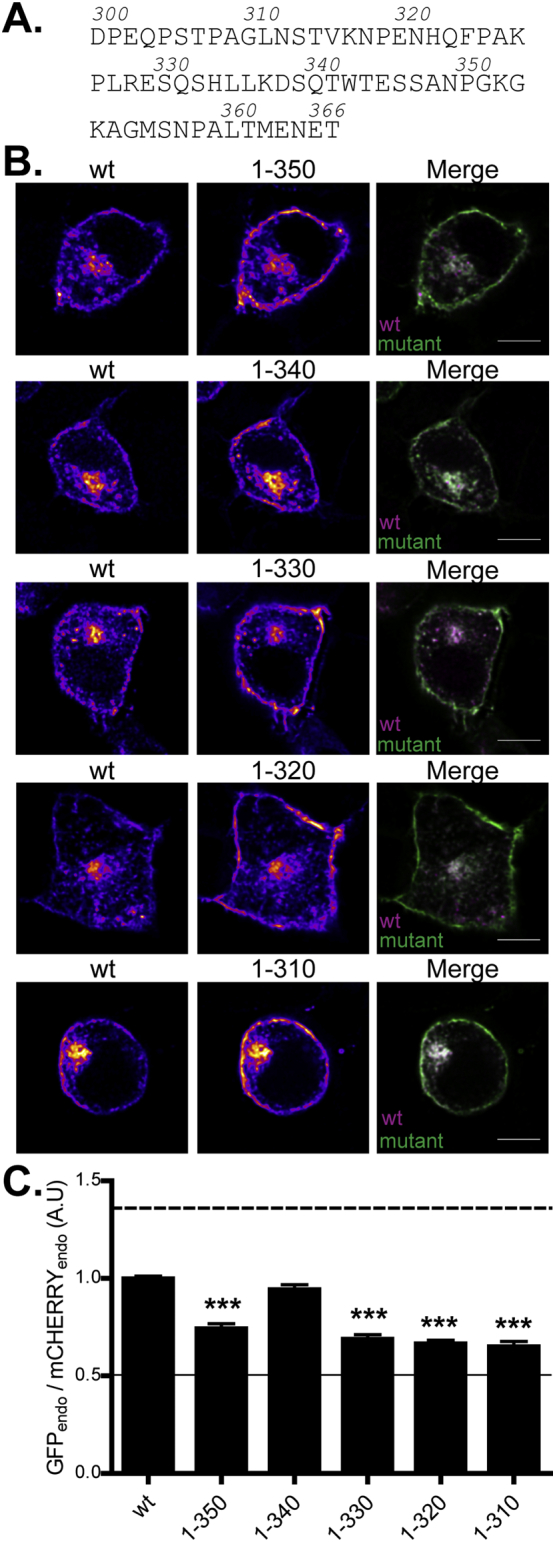
Localisation of zDHHC2 truncation constructs. (A) Amino acid sequence of the C-terminal cytosolic tail of mouse zDHHC2. The numbers indicate the position of the amino acid upstream of the inserted STOP codon, allowing for the expression of zDHHC2 truncated proteins. (B) Representative confocal images of PC12 cells co-expressing the wt mCHERRY-zDHHC2 protein (left panel) and the GFP-tagged truncation mutants of zDHHC2 (middle panel). Both are represented in FIRE pseudocolour. The right panel is the merge of the left and middle panels, with the wt mCHERRY-zDHHC2 presented in Magenta and GFP-tagged proteins in Green. Scale bar represents 5 μm. (C) Quantification of the endosomal depletion of the truncation mutants. The fluorescence intensity value of the recycling endosome compartment (endo) as a fraction of the whole cell fluorescence was calculated as described in material and methods for GFP constructs relative to the wt zDHHC2 mCHERRY-tagged protein (n = 14 wt zDHHC2 cells, 16 (1–350) cells, 15 (1–340) cells, 14 (1–330) cells, 15 (1–320) cells and 17 (1–310) cells). Statistical analysis (ANOVA) shows a significant difference in the localisation of several truncation mutants compared to wt zDHHC2 (⁎⁎⁎, p ≤ 0.001). The dashed and filled lines represent the upper and lower ratios of endosomal enrichment that were calculated in Fig. 1. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
