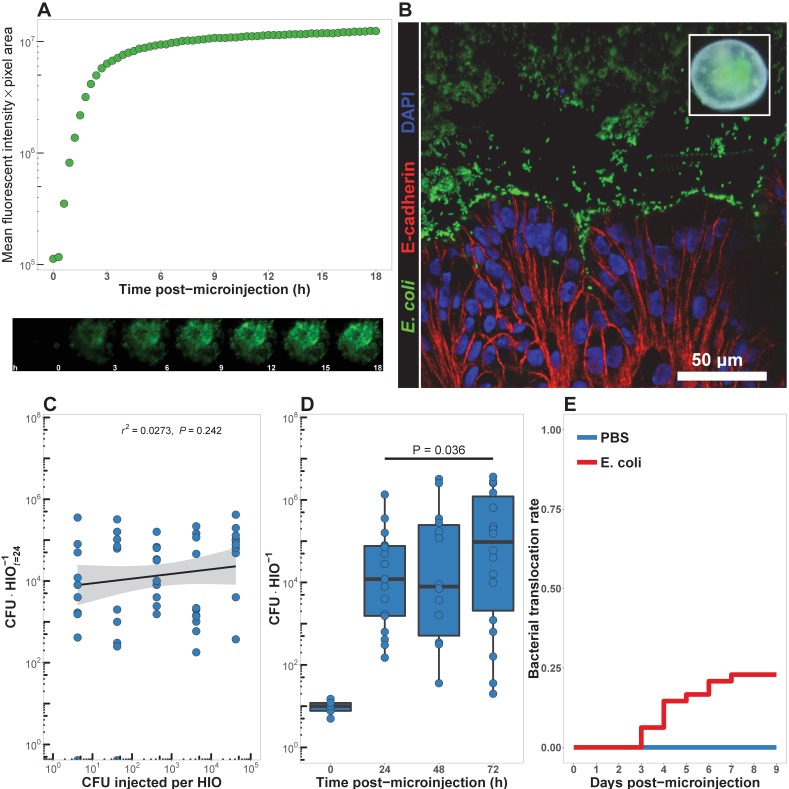
Figure 1. HIOs can be stably associated with non-pathogenic E. coli.
(A) Mean fluorescent intensity of a human intestinal organoid (HIO) containing live GFPE. coli str. ECOR2. The lower panels show representative images from the time series. Representative of three independent experiments. Video 1 is an animation corresponding to this dataset. (B) Confocal micrograph of the HIO epithelium (E-cadherin) in direct association with GFP+ E. coli at 48 hr post-microinjection with 104 live E. coli. 60X magnification. (C) Luminal CFU per HIO E. coli at 24 hr post-microinjection relative to the injected concentration of 5 × 10-1 to 5 × 105 CFU per HIO at the start of the experiment. N = 10 biological replicates per E. coli dose. The r and P value shown in the figure represent the results of a linear regression analysis of the relationship between the 24 hr change in CFU/HIO and the initial number of CFU injected. (D) Luminal CFU per HIO at 0–72 hr following microinjection with 10 CFU E. coli per HIO. N = 13–17 replicate HIOs per time point. The p-value represents the results of a two-tailed Student’s t-test comparing the two conditions indicated. (E) Daily proportion of HIO cultures with no culturable E. coli in the external media following E. coli microinjection (N = 48) or PBS microinjection (N = 8).