Figure 4.
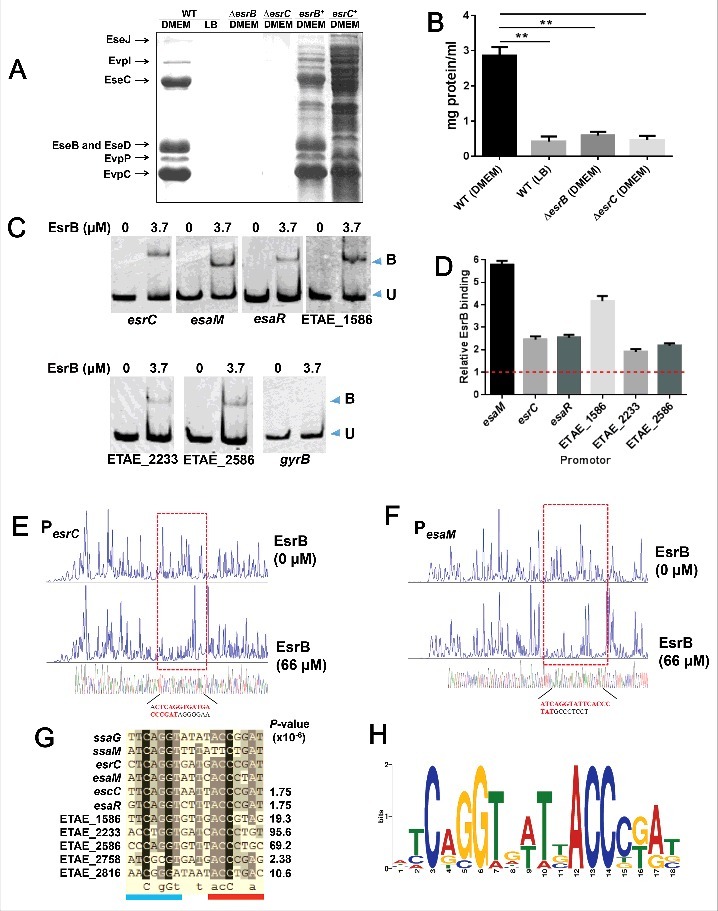
EsrB binds directly to a specific DNA motif to control the expression of T3SS and other genes. (A) ECP profiles of EIB202 WT and ΔesrB were detected by SDS-PAGE. Strains were cultured in LB and DMEM, respectively, at 30°C for 24 h without shaking. (B) Total protein secreted when the strains were grown in LB or DMEM conditions. The secreted proteins were quantitatively assayed against controls consisting of DMEM. **, P < 0.01 based on one-way ANOVA. (C) EMSA of the indicated promoter regions using purified EsrB. The amounts of EsrB protein used are indicated; 20 ng of each Cy5-labeled probe was added to the EMSA mixtures. The specificity of the shifts was verified by adding a 10-fold excess of nonspecific competitor DNA poly(dI·dC) to the EMSA mixtures. Bound (B) and unbound (U) DNA bands are indicated. (D) ChIP-qPCR analysis was used to determine the binding of EsrB to target promoter regions in E. piscicida. The results shown are normalized to the expression of the control gene gyrB and the results from the sample without EsrB, which was arbitrarily set as 1. The results were calculated using the ΔΔCT method. (E-F) Footprinting analysis of EsrB binding to a binding site in the esrC and esaM promoter. Electropherograms of a DNase I digest of the promoter probes (200 ng) after incubation with 0 or 66 µM EsrB are shown. The respective nucleotide sequences that were protected by EsrB are indicated below, and the specific binding motifs are highlighted. (G) Alignment of the EsrB-binding motifs from the promoter regions of EsrB-controlled genes as revealed by RNA-seq analysis. The established binding motifs from ssaG and ssaM in Salmonella enterica are also shown. P-values are shown for the genes identified with FIMO. (H) The EsrB binding motif derived from the binding sequences (G) generated by the MEME tool. The height of each letter represents the relative frequency of each base at the given position in the consensus sequence.
