Figure 1.
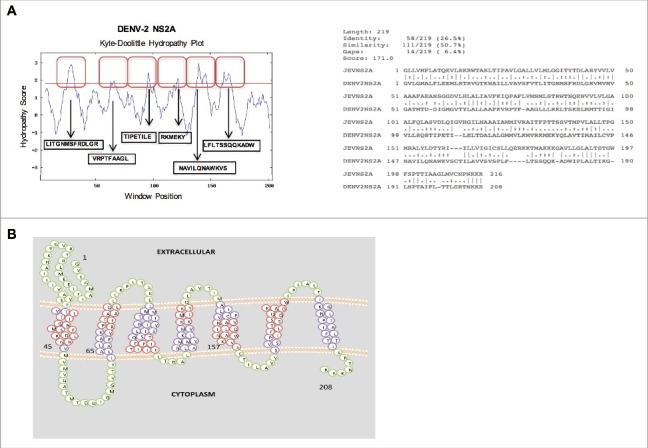
In silico analysis of the amino acid sequence (A) (Above) Hydropathy plots of the NS2A protein of Dengue virus (DENV), as generated via the Kyte-Doolittle method and DAS TM prediction algorithm using SOSUI software. Red boxes indicate the highly hydrophobic transmembrane regions of NS2A. (Below) Alignment of the amino acid sequences of NS2A from DENV and Japanese encephalitis virus (JEV), as generated using ClustalW software. Identical (*), similar (.), and low-similarity (:) amino acids were observed. (B) Proposed model of NS2A topology generated using SOCS MEMSAT software. Trans-membrane segments are shown in purple, while the green color indicates inter-transmembrane regions of NS2A. Regions highlighted in red denote highly hydrophobic, possible membrane interacting, transmembrane segments.
