ABSTRACT
Up to now, very small protein-coding genes have remained unrecognized in sequenced genomes. We identified an mRNA of 165 nucleotides (nt), which is conserved in Bradyrhizobiaceae and encodes a polypeptide with 14 amino acid residues (aa). The small mRNA harboring a unique Shine-Dalgarno sequence (SD) with a length of 17 nt was localized predominantly in the ribosome-containing P100 fraction of Bradyrhizobium japonicum USDA 110. Strong interaction between the mRNA and 30S ribosomal subunits was demonstrated by their co-sedimentation in sucrose density gradient. Using translational fusions with egfp, we detected weak translation and found that it is impeded by both the extended SD and the GTG start codon (instead of ATG). Biophysical characterization (CD- and NMR-spectroscopy) showed that synthesized polypeptide remained unstructured in physiological puffer. Replacement of the start codon by a stop codon increased the stability of the transcript, strongly suggesting additional posttranscriptional regulation at the ribosome. Therefore, the small gene was named rreB (ribosome-regulated expression in Bradyrhizobiaceae). Assuming that the unique ribosome binding site (RBS) is a hallmark of rreB homologs or similarly regulated genes, we looked for similar putative RBS in bacterial genomes and detected regions with at least 16 nt complementarity to the 3′-end of 16S rRNA upstream of sORFs in Caulobacterales, Rhizobiales, Rhodobacterales and Rhodospirillales. In the Rhodobacter/Roseobacter lineage of α-proteobacteria the corresponding gene (rreR) is conserved and encodes an 18 aa protein. This shows how specific RBS features can be used to identify new genes with presumably similar control of expression at the RNA level.
Keywords: α-proteobacteria; Bradyrhizobium; ribosomal binding site; ribosome; RNA stability; Shine-Dalgarno; small gene; small protein; sORF; translation initiation region
Introduction
Deep sequencing of bacterial transcriptomes revealed many non-annotated small transcripts, some of which are probably regulatory non-coding RNAs (sRNAs), small mRNAs, or RNAs with dual functions.1-4 Most bacterial mRNAs harbor Shine-Dalgarno sequences (SDs) in their 5′-untranslated regions (5′-UTRs), which mediate the interaction of the mRNAs with the 30S ribosomal subunit.5,6 Thus, the identification of canonical SDs should be helpful for identification of small functional open reading frames (sORFs) that are ignored in genome annotations.3,7
A canonical SD is located 5 to 9 nucleotides (nt) upstream of the translation initiation codon and has the sequence 5′-AGGAGG-3′ that is perfectly complementary to the 3′-end of 16S rRNA.5,6 However, most SDs are not perfectly complementary to 16S rRNA and canonical 6-meric SDs are strongly underrepresented in bacterial genomes.6 Moreover, SDs longer than 6 nt with perfect complementarity to 16S rRNA have rarely been described. An extended SD consisting of 8 nt was shown to enable translation of a chloroplast mRNA from a downstream GUG start codon instead of an upstream AUG codon.8 Further, artificially extended SDs of 8 and 10 nt resulted in lower translation efficiency in E. coli, probably due to extraordinary strong binding to the anti-Shine-Dalgarno (aSD) in 16S rRNA, thus impeding transition from initiation to elongation of translation.9 However, in another study with E. coli an 8 nt SD led to higher translation efficiency than SDs of 6 nt or 4 nt.10
Identification of small protein-encoding genes has remained a challenge, but examples of small proteins that fulfill important functions underline the importance of this research field.3,7,11 Among others, small proteins influence resistance to antibiotics, regulate bacterial communication, or act as non-secreted toxins contributing to formation of persister cells.12-16 Recently we re-annotated the genome of the Bradyrhizobium japonicum USDA 110, a soil-dwelling bacterium capable of fixing molecular nitrogen in symbiosis with several legume plants including soybean.17,18 This re-annotation added 1,391 potential ORFs to the 8,317 originally annotated genes.19,20 Although sORFs shorter than 30 codons were not annotated, it is conceivable that such sORFs remain to be identified and that some of them are encoded by putative sRNAs. Many transcription start sites (TSSs) of orphan genes that may correspond to sRNAs or small mRNAs were recently mapped in the genome of B. japonicum USDA 110.19
In this work, we analyzed a small RNA in B. japonicum USDA 110 and found that it is an mRNA conserved in Bradyrhizobiaceae. It encodes a polypeptide chain comprising 14 amino acid residues (aa) and harbors an extremely large SD of 17 nt, which extends to a GTG start codon. The polypeptide chain, synthesized by solid-phase chemical synthesis, does not adopt a persistent secondary or tertiary structure as judged by CD- and NMR-spectroscopic analyses. We show that the unique ribosome binding site (RBS) is responsible for strong interaction with ribosomes and low translation efficiency. Furthermore, our data indicate ribosome-dependent degradation of the small mRNA. Thus, the corresponding gene was named rreB (ribosome-regulated expression in Bradyrhizobiaceae). Small genes with similar RBS, which are probably regulated at the RNA level in a similar way, were also found in other α-proteobacteria.
Experimental procedures
Cultivation methods and cloning procedures
B. japonicum 110spc4, a spectinomycin-resistant derivative of B. japonicum USDA 110 (recently re-named to Bradyrhizobium diazoefficiens), was grown in PSY medium in the presence of spectinomycin at a concentration of 100 μg ml−1 at 30°C.21-23 Escherichia coli JM109 and E. coli S17-1 were cultivated in LB medium.24,25 Standard cloning procedures were used.26 Suitable constructs for ectopic expression in B. japonicum were cloned in pME3535XhoI or its derivatives, cleaved out with EcoRI and XhoI restriction endonucleases and re-cloned into the broad host range plasmid pRK290XhoI resulting in pJH-plasmids (see Table S1).27,28 Additionally, the chromosome integration plasmid pRJPaph-gfp_a1 was used.29 The gfp gene of this plasmid was cleaved out with SpeI and KpnI and was replaced by a sequence containing BamHI, PstI and SphI restriction sites. The resulting plasmid pRJPaph-MCS was used to clone rreB derivatives. Plasmids were transferred from E. coli S17-1 to B. japonicum 110spc4 by biparental conjugation.25 Used plasmids and oligonucleotides are listed in Tables S1 and S2, respectively.
RNA isolation and analysis
Total RNA was isolated with hot-phenol.30 Soybean nodules were kindly provided by H.M. Fischer (ETH Zürich). RNA separation in urea-containing 10 % polyacrylamide gels, staining with ethidium bromide, semidry blotting, hybridization with radioactively labeled oligonucleotide probes, re-hybridization, signal detection and quantification were performed as described.31 Oligonucleotides used for hybridization are listed in Table S2. RNA stability measurements after stop of transcription with rifampicin were performed as previously described.32
Fractionation of cell-free extracts
B. japonicum cells were harvested by centrifugation at 13,000 g at 4°C, re-suspended in a buffer containing 20 mM HEPES-KOH (pH 7.8), 10 mM MgCl2 and 100 mM NaCl, and lysed by sonication. S100 and P100 fractions were obtained after ultracentrifugation at 100,000 g. RNA isolated from the S100 fraction and the P100 fraction was dissolved in the same volume and same volumes were analyzed by Northern blot hybridization. The P100 fraction was resuspended in the same buffer and further fractionated in 10 ml 5% to 40% sucrose density gradient containing 3 mM MgCl2 at 75,000 g and 4°C for 16 h. Fractions (0.5 ml) were harvested from the top and absorption at 260 nm was measured to localize the 30S and 50S ribosomal subunits.33
Western blot analysis
Exponentially growing B. japonicum cells were harvested and adjusted to an OD600 of 10 in SDS-containing loading buffer. Cells were lysed by incubation at 100°C for 5 min, proteins were separated by SDS-PAGE and blotted onto a nitrocellulose membrane.26 If necessary, cell suspensions were diluted tenfold prior to lysis. Detection was performed with GFP-specific antibodies (Clontech), Anti-Mouse IgG-Alkaline Phosphatase (Sigma) and CDP-Star (Roche). Signals were visualized using a chemiluminescence imager (Fusion SL4, Vilber).
Flow cytometry and cell sorting
Flow cytometry analysis and cell sorting experiments were performed with a FACSAria II flow cytometer (Becton Dickinson, Heidelberg, Germany). For excitation, a blue solid-state laser with a wavelength of 488 nm was used. Forward-scatter characteristics (FSC) and side-scatter characteristics (SSC) were detected as small-angle and orthogonal scatter of the 488-nm laser. For the detection of EGFP fluorescence a 502 nm long-pass and a 530/30 nm band-pass filter set was used. The FACS-Diva software 6.0 was used for measurement and data recording. Tracking beads labeled with a mixture of fluorochromes (BD, Heidelberg, Germany) were used for the cytometer set-up and performance tracking. Noise was removed by thresholding on FSC and SSC, while cell size was discriminated over the intensity of the FSC signal. Four-way purity was used as the precision mode for cell sorting with a threshold rate of up to 8000 events/sec. Data were analyzed using FlowJo V10 (Tree Star, Ashland, USA).
Peptide synthesis, CD and NMR analyses
The small polypeptides rreB from B. japonicum USDA 110 (14 aa) and rreR from Dinoroseobacter shibae DFL 12 (18 aa) were synthesized using standard Fmoc-chemistry and purified by reversed-phase HPLC. The purity of the synthesized products characterized by matrix-assisted laser desorption/Ionization (MALDI) mass analysis was analytical HPLC and greater 95% for both peptides. Secondary structure analysis was performed by 1D 1H-NMR experiments and circular dichroism (CD) spectroscopy (see Supplementary Methods).
Bioinformatic analyses
Homologs of rreB were identified by BLASTN and were used for multiple sequence alignments and secondary structure prediction.34,35 Clustal W was also used for multiple sequence alignments.36 Phyre2 was used for prediction of structures of proteins.37
Results
A small mRNA with extraordinarily long Shine-Dalgarno sequence in Bradyrhizobiaceae
Based on existing transcriptome data and previously mapped TSSs, promoters, and terminators, we identified a sRNA with a length of 165 nt, a TSS located at genomic position 6,046,962 and a putative RpoD-like promoter (Fig. 1A).19 Northern blot analysis confirmed this transcript in exponentially growing and stationary free-living B. japonicum cells, and in symbiosis with soybean (Fig. 1B). In addition to the expected band of approximately 165 nt, the Northern blot analysis revealed a putative degradation product of approximately 120 nt present only in symbiosis and a putative degradation product of approximately 60 nt under all tested conditions (Fig. 1B).
Figure 1.
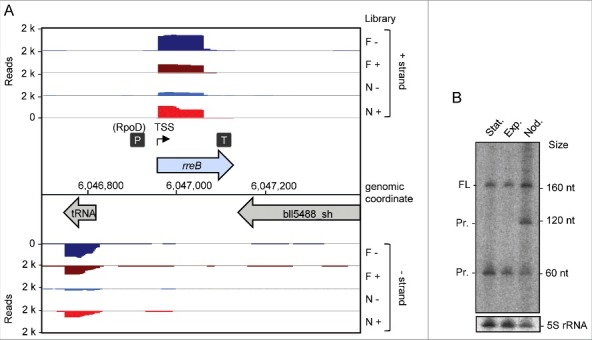
Detection of the small mRNA rreB. (A) Genomic context of rreB and cDNA reads, which originate from a previous differential RNA-seq analysis.19 RNA was isolated from exponentially growing, free-living cells in liquid cultures (F) and from nodules (N). RNA samples were treated (+) or not treated (−) with terminal exonuclease (TEX), which degrades 5′-monophosphorylated (processed) transcripts. The scale of each library is indicated (Reads). Flexed thin black arrow, mapped TSS; dark gray boxes with P and T, mapped putative promoter and terminator, respectively.19 The gene rreB is located between Bjat37 encoding tRNA-Gly and bll5488_sh encoding a 2-component hybrid sensor and regulator. (B) Northern blot hybridization for detection of the sRNA rreB using a probe complementary to the sORF (see Fig. 2A). Total RNA isolated from liquid cultures grown to the exponential growth phase (Exp), the stationary phase (Stat) and from soybean nodules (Nod) was used. On the right side, the positions of the marker RNAs (160 nt, 120 nt and 60 nt corresponding to 6S rRNA, 5S rRNA and a fragment detected by the 6S RNA-specific probe, respectively30), are given. The full-length transcript (FL) and putative degradation products (Pr.) were detected. 5S rRNA was used as loading control.
The small RNA contains a sORF encoding a polypeptide chain (referred to here as small protein) of 14 aa. Interestingly, the SD upstream of the sORF is a part of a 17 nt region showing perfect complementarity to the 3′-end of 16S rRNA (17 nt complemetarity-region or 17c-region; Fig. 2A). The complementarity region encompasses bases upstream and downstream of the canonical SD and includes the first base of the GTG start codon. Such an unprecedented SD extension should lead to a very strong binding of the small mRNA to 16S rRNA in the 30S ribosomal subunit and may negatively influence or even abolish translation of the small protein.9 BLASTN analyses revealed that the small mRNA is conserved in the family Bradyrhizobiaceae (Fig. 2A and Fig. S1). The corresponding gene was named rreB (ribosome-regulated expression in Bradyrhizobiaceae; see below).
Figure 2.
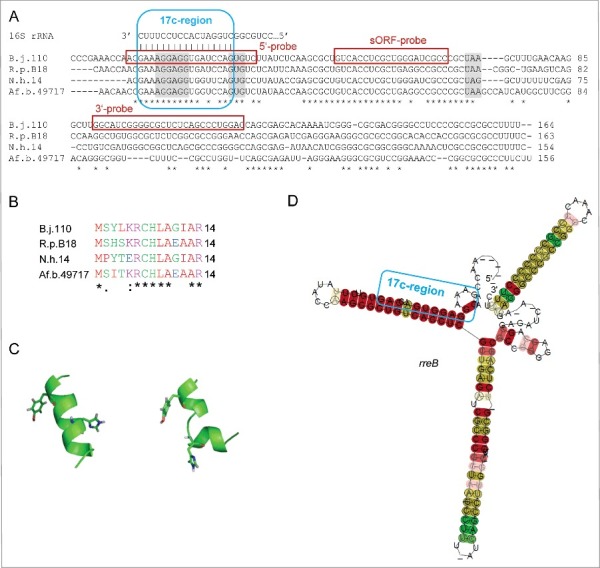
The conserved small RNA rreB contains a sORF and shows extended complementarity to the 3′-end of 16S rRNA. (A) Multiple sequence alignment of rreB mRNA and its homologs in 4 genera of Bradyrhizobiaceae. Asterisks mark invariant positions. The canonical SD sequence 5′-AGGAGG-3′, the GUG start codon and UAA or UAG stop codons are highlighted. The sequence of the 3′-end of 16S rRNA is shown on the top. The 17 nt region of perfect complementarity between rreB and 16S rRNA (17c-region) is framed in blue. Sequences targeted by 3 different probes in Northern blot hybridizations are framed in red (see Fig. 3 below). B.j.110, B. japonicum USDA 110; R.p. B18, Rhodopseudomonas palustris BisB18; N.h. 14, Nitrobacter hamburgensis X14; Af. b. 49717, Afipia broomeae ATCC 49717. An extended alignment is shown in Fig. S1. (B) Multiple sequence alignment of the small proteins RreB encoded by the sORFs presented in A). An extended alignment is shown in Fig. S1. (C) Predicted spatial structure of RreB from B. japonicum USDA 110. Aromatic amino acid residues are marked. (D) Predicted RNA secondary structure of the rreB homologs shown in A). The LocARNA color annotation shows the conservation of base pairs - highly reliable prediction is indicated by the red color.36 Less conserved basepairs are indicated by grey letters.
The small protein encoded by rreB is conserved (Fig. 2B and Fig. S1) and has predicted α-helix and random coil regions (Fig. 2C). It was synthesized by standard Fmoc-chemistry and its structure was analyzed by CD and 1D-NMR spectroscopy (Figs. S2 to S5). However, the small protein RreB remained unstructured under all tested conditions, suggesting that it may adopt a structure only upon binding to interaction partner(s).
The small mRNA rreB binds to the 30S ribosomal subunit
The presence of rreB homologs in several Bradyrhizobiaceae genera allowed prediction of a conserved secondary structure of the small mRNA. The prediction suggests that with exception of the 5 most upstream nucleotides, the 17c-region and the GTG start codon are involved in a double-stranded structure (Fig. 2C). The putative formation of double-stranded mRNA structure raised the question whether the 5′-UTR of rreB is accessible for interaction with ribosomes. To test whether rreB is bound to ribosomes, lysates of exponentially growing B. japonicum cultures were fractionated by ultracentrifugation at 100.000 g in supernatant (S100) fraction containing the soluble components of the cell and pellet (P100) fraction containing membranes and ribosomes. RNA from both fractions was analyzed by Northern blot hybridization along with total RNA.
The Northern blot analysis was performed with 3 probes directed to the 17c-region, the sORF (the same probe was used in Fig. 1B) and the 3′-UTR, respectively (Fig. 3A). As expected, with the sORF probe the 165 nt full-length rreB mRNA and the 60 nt degradation product were detected in total RNA. In contrast, the 60 nt band was not detected with probes directed to the 5′- and 3′-UTR, respectively (Fig. 3A). After 100.000 g fractionation, the 60 nt band was detected predominantly in the S100 fraction, while the 165 nt band was mainly in the P100 fraction (Fig. 3A). As control for successful fractionation we used tRNA-Arg (Bjat49) which is not expected to be fully associated with ribosomes and was mainly in the S100 fraction, and 5S rRNA, which as part of the 50S ribosomal subunit was mainly in the P100 fraction (Fig. 3B). Thus, in contrast to the 60 nt processing product, most of the full-length rreB mRNA seemed to be associated with ribosomes.
Figure 3.
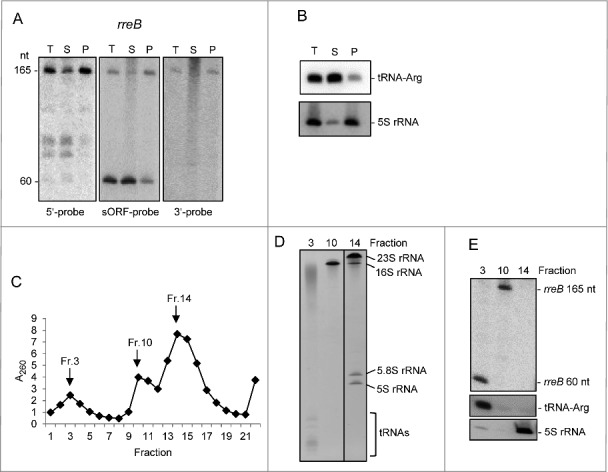
The small mRNA rreB is in the insoluble fraction and is strongly bound to the 30S ribosomal subunit in B. japonicum. (A) Northern blot analysis of total RNA (T) and RNA isolated from the P100 (P) and S100 (S) fraction using probes directed against the 5′-UTR (5′-probe), the sORF (ORF-probe) and the 3′-UTR (3′-probe) of rreB as indicated; the same membrane was re-hybridized. Main bands of approximately 165 nt and 60 nt were detected (marked at the left side). Their lengths were estimated by hybridization of the membrane with probes directed against the length standards 6S RNA (160 nt), 5S rRNA (120 nt) and tRNA-Arg (79 nt) (not shown). (B) Northern blot analysis of tRNA-Arg and 5S rRNA. The membrane shown in A) was re-hybridized. (C) Separation of the P100 fraction through a 5% to 40% sucrose density gradient. The absorption of the gradient fractions at 260 nm is shown. Fractions which were further analyzed are marked with arrows. (D) Analysis of RNA isolated from fractions 3, 10 and 14 of the sucrose density gradient by electrophoresis. RNA was separated in urea-containing 10% polyacrylamide gel and stained with ethidium bromide (shown is a negative image). Distinct bands corresponding to the large 23S rRNA fragment (23S rRNA), 16S rRNA, the 5.8S rRNA-like short 5′-fragment of 23S rRNA, 5S rRNA and tRNAs are indicated. Fragmentation of 23S rRNA in Bradyrhizobium japinicum was described previously.40,41 (E) Northern blot hybridization of RNA from the indicated sucrose density gradient fractions with probes directed against the sORF of rreB, tRNA-Arg and 5S rRNA.
The P100 fraction was further analyzed by ultracentrifugation in sucrose density gradients in which the ribosomal subunits were essentially separated (Fig. 3C and 3D). Twenty-two fractions were collected from the top of the gradient and RNA was isolated from the peak fractions 3 (containing mRNAs and tRNAs), 10 (containing the 30S ribosomal subunit) and 14 (containing mainly the 50S ribosomal subunit; see Fig. 3C and 3D). Using Northern blot hybridization we detected the 165 nt rreB band in fraction 10 and the 60 nt rreB band in fraction 3 (Fig. 3E). As expected, the tRNA-Arg was in fraction 3 and 5S rRNA in fraction 14 (Fig. 3E). These results suggest that the 60 nt degradation product easily dissociates from the ribosomes, and show that full-length rreB mRNA is strongly bound to the 30S ribosomal subunit.
The 5′-extension of the SD is crucial for predominant localization of rreB at ribosomes
To analyze the impact of the 17c-region on the binding to ribosomes, we ectopically overexpressed 3 5′-truncated rreB mRNA derivatives (Fig. 4A): 1) 17c-rreB lacking the first 8 nt of wild type rreB but still having the 17 nt complementary to 16S rRNA (compare Fig. 4A to Fig. 2A), 2) 14c-rreB starting with the full canonical SD and having 14 nt complementary to 16S rRNA, and 3) 8c-rreB lacking the canonical SD and having 8 nt complementary to 16S rRNA. When compared with endogenous rreB, the overexpression of the truncated rreB derivatives was strong in B. japonicum, ranging from 20-fold for 17c-rreB to 120-fold for 8c-rreB, as determined by Northern blot hybridization (Fig. 4B). The overexpressing strains were subjected to S100 and P100 fractionation followed by Northern blot hybridization, showing that 17c-rreB was predominantly in the P100 fraction and the other 2 rreB derivatives with shortened region of complementarity to 16S rRNA were mainly in the S100 fraction (Fig. 4C). The strong difference in the localization of 17c-rreB and 14c-rreB shows that the most upstream nucleotides of the 17c-region are crucial for the predominant localization of rreB in P100 and thus for binding to ribosomes in B. japonicum. When analyzed in E. coli, the rreB derivatives accumulated to very low levels and were mainly in the S100 fraction (Fig. S6). To test whether 20-fold overproduction of 17c-rreB mRNA that tightly binds to ribosomes influences B. japonicum growth, growth curves of the overexpressing strain and the EVC were compared but no difference was found (not shown).
Figure 4.
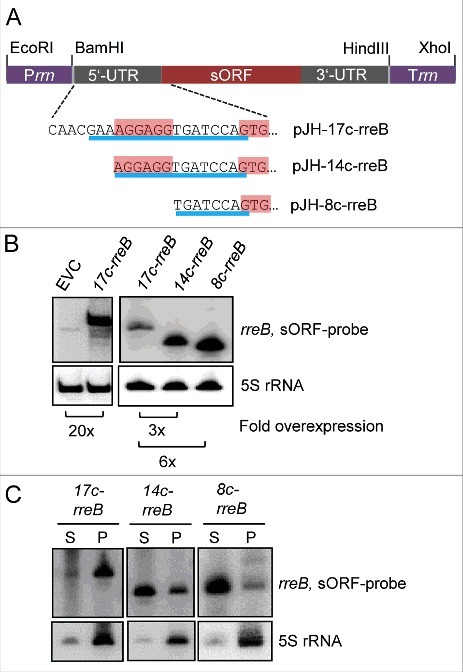
The 5′-nucleotides of the 17c-region are crucial for the predominant localization of rreB in the P100 fraction. (A) Schematic representation of the used constructs. The 5′-UTRs and GTG start codons of the rreB derivatives overexpressed from plasmids pJH-17c-rreB, pJH-14c-rreB, and pJH-8c-rreB are shown. The region of complementarity to 16S rRNA is underlined in blue. The canonical SD and the GTG start codon are highlighted. Prrn, rRNA (rrn) promoter; Trrn, rrn terminator. (B) Northern blot hybridization of total RNA of B. japonicum strains overproducing the indicated rreB derivatives. EVC, empty vector control. (C) Northern blot hybridization of RNA isolated from the P100 (P) and S100 (S) fractions of strains overproducing the indicated rreB derivatives. (B) and (C) shown results from representative experiments. In each case, 3 independent biological experiments with similar results were performed. Probes directed against rreB and the 5S rRNA control were used.
The RBS of rreB suppresses translation
The strong binding of rreB to 16S rRNA may negatively influence translation.9 To test whether translation of rreB takes place in B. japonicum, we cloned rreB-egfp translational fusions. For this purpose first a control plasmid pJH-F2 was created, which contains the B. japonicum rrn promoter, a 5′-UTR with a standard SD and an ATG start codon (standard RBS) from the bacterial overexpressing vector pQE30 (Qiagen), in frame egfp, and the B. japonicum rrn terminator. Plasmid pJH-F2 leads to strong egfp expression in B. japonicum (Fig. 5B, lane 2). Suitable restriction sites were included in pJH-F2 in order to enable construction of egfp translational fusions by replacement of the standard RBS (Fig. 5A).
Figure 5.
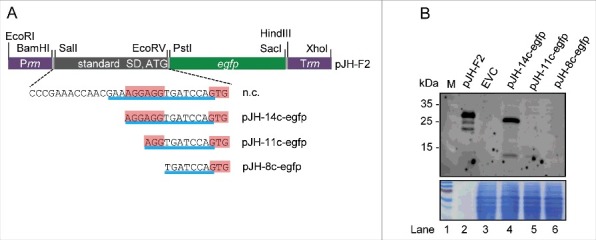
Analysis of the influence of truncated rreB 5′-UTRs on EGFP translation. (A) Schematic representation of plasmid pJH-F2 and of its derivatives pJH-14c-egfp, pJH-11c-egfp, and pJH-8c-egfp, which contain rreB-egfp translational fusions. The standard SD and the ATG start codon of pJH-F2 were replaced by the indicated rreB sequences. n.c., cloning of a construct with the full rreB 5′-UTR without the rreB sORF failed. Prrn and Trrn, see Fig. 4A. (B) Western blot analysis with GFP-specific antibodies of strains containing the indicated constructs (see A). EVC, strain containing plasmid pJH-O1. The bottom panel shows a Coomassie Blue stained SDS-polyacrylamide gel after electrophoresis visualizing the loaded protein amounts. Migration of marker proteins (M) in the gel is indicated at the left side in kDa.
To test the influence of the 17c-region on translation, we aimed to replace the standard RBS by the full rreB 5′-UTR and the GTG start codon, but we failed to clone such a construct (marked with n.c. in Fig. 5A). Therefore, we cloned truncated derivatives with 14, 11 and 8 nt complementarity to 16S rRNA (pJH-14c-egfp, pJH-11c-egfp and pJH-8c-egfp; Fig. 5A). EGFP production in B. japonicum cells was monitored by Western blot analysis with GFP-specific antibodies (Fig. 5B). Signals corresponding to EGFP (33 kDa) were detected only in lysates with pJH-14c-egfp (lane 4) and the positive control pJH-F2 (lane 2). The EGFP signal in lane 4 was much weaker than that in lane 2 (consider the bottom panel which shows the loaded protein amounts in Fig. 5B). Thus, the RBS with the 14c-region supports weak translation, while the truncated 11c- and 8c-derivatives (lanes 5 and 6 in Fig. 5B) do not support it. Considering the strong overproduction of 14c-rreB mRNA (Fig. 4B) and the fact that its truncated 5′-UTR supports translation (Fig. 5B), we tested whether the increased RreB level influences growth. However, we did not found difference between the growth of B. japonicum (pJH-14c-rreB) and the EVC (not shown).
To analyze the impact of the full-length 5′-UTR of rreB on translation, we cloned the 5′-UTR together with the sORF in a translational fusion with egfp. Western blot analysis of cultures from cells containing the resulting plasmid pJH-17c-rreB-egfp revealed the presence of very low amounts of degraded EGFP (Fig. S7). Therefore we re-cloned the 17c-rreB-egfp construct to obtain a chromosome-integrating plasmid pRJ-17c-rreB-egfp, in which, the rreB-egfp translational fusion is under the control of a constitutive Paph promoter (Fig. 6A).29 As a corresponding control pRJ-sSD-rreB-egfp was created, in which the standard RBS precedes the rreB-egfp fusion (Fig. 6A). The Western blot analysis in Fig. 6B shows that cells harboring pRJ-17c-rreB-egfp the fusion protein RreB-EGFP. The RreB-EGFP band from those cells was very weak when compared with the positive control with pRJ-sSD-rreB-egfp (compare lanes 4 and 5 in Fig. 6B considering the loaded protein amounts), in line with weak RreB-EGFP translation due to the 17c-region.
Figure 6.
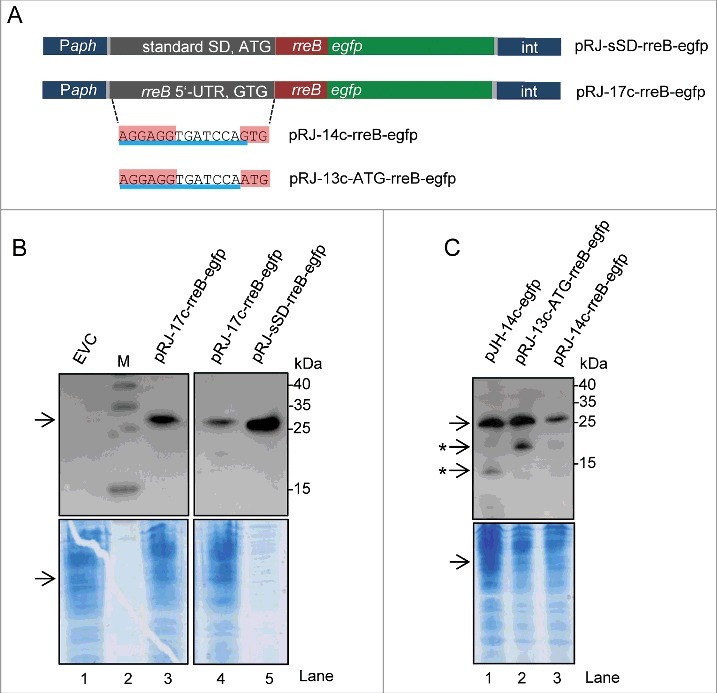
The mRNA rreB is weakly translated in B. japonicum. (A) Schematic representation of the used constructs cloned between a Paph promoter and a sequence for integration of the plasmids into the chromosome (int).29 The rreB-egfp fusion is preceded either by a standard SD and an ATG start codon (pRJ-sSD-rreB-egfp) or by the full-length 5′-UTR of rreB with the 17c-region and the GTG start codon (pRJ-17c-rreB-egfp). Further modifications of the 5′-UTR of rreB resulting in pRJ-14c-rreB-egfp and pRJ-13c-ATG-rreB-egfp are indicated on the bottom. (B) and (C) Western blot analysis with GFP-specific antibodies of strains containing the indicated constructs (see A). EVC, plasmid pJPaph-MCS was integrated into the chromosome. Schematic representation of of pJH-14c-GTG-egfp is given in Fig. 5A. The bottom panel shows a Coomassie Blue stained SDS-polyacrylamide gel after electrophoresis visualizing the loaded protein amounts. Migration of marker proteins (M) is indicated (in kDa). Arrows indicate the position of the RreB-EGFP band; arrows with asterisks indicate putative degradation products.
B. japonicum cells with the constructs shown in Fig. 6A were also examined under the fluorescence microscope. While 90% of the positive control cells harboring pRJ-sSD-rreB-egfp were fluorescent, in cultures with pRJ-17c-rreB-egfp there were almost no fluorescent cells (not shown). To test whether the fusion protein RreB-EGFP accumulates only in a small sub-population, the strain harboring pRJ-17c-rreB-egfp was analyzed additionally by flow cytometry. For thresholding we used an empty vector control (EVC) strain containing chromosomally integrated pRJPaph-MCS as reference and for the adjustment of the gating strategy. No significant difference was detected between cultures of the EVC and the pRJ-17c-rreB-egfp containing strain, both showing 0.03% fluorescent cells (Fig. S8). The fluorescence of those cells was weak and probably represents auto-fluorescence. Single fluorescent and non-fluorescent cells were spotted onto agar plates using FACS. RreB-EGFP was detected by Western blot analysis with GFP-specific antibodies in all cultures originating from single cell isolates with pRJ-17c-rreB-egfp and the signals were of similar intensities (Fig. S8). Altogether, these results suggest that the level of the fusion protein RreB-EGFP is very low in the cells and therefore its fluorescence is under the limit of detection.
To analyze the impact of the GTG codon on rreB translation, we constructed a pRJ-14c-rreB-egfp plasmid with the GTG start codon and a mutated derivative pRJ-13c-ATG-rreB-egfp in which the GTG codon was replaced by ATG (Fig. 6A). Western blot analysis revealed that the ATG codon mediates higher translation efficiency than the GTG codon (compare lanes 2 and 3 in Fig. 6C).
Based on the above data, we conclude that the sORF rreB is translated, but its translation is very weak due to the extremely extended SD and the GTG codon.
The start codon is important for rreB turn-over
In the course of our analyses, we constructed a GUG/UAG mutant of the overexpressed 17c-rreB RNA (see Fig. 4A), in which the start codon GUG is replaced by the stop codon UAG. This derivative named 16c-rreB(GUG/UAG) was localized predominantly in the P100 fraction (Fig. 7A), suggesting that it binds to ribosomes with similar efficiency like 17c-rreB or endogenous rreB. However, the 16c-rreB(GUG/UAG) transcript accumulated to higher levels in the cells than 17c-rreB (not shown). Since both RNAs are transcribed from the rrn promoter, this observation suggested that 16c-rreB(GUG/UAG) is more stable than 17c-rreB, pointing to a function of the ribosome in rreB degradation. We performed stability measurements and found that the GUG/UAG mutation significantly increased the half-life of the transcript from 10 ± 2 min to 37 ± 13 min (Fig. 7B and 7C). Thus, either formation of 30S initiation complex or 70S ribosome are involved in degradation of 17c-rreB, suggesting an additional regulatory role of the ribosome in rreB expression.
Figure 7.
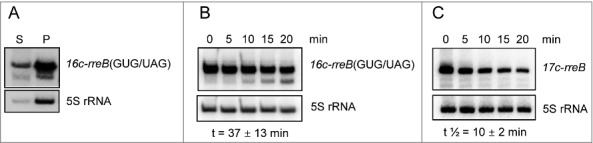
The rreB start codon is important for the turnover of the small mRNA. Northern blots hybridized with probes directed against the rreB sORF and the control 5S rRNA are shown. (A) The 16c-rreB(GUG/UAG) transcript is localized predominantly in the P100 fraction. (B) and (C) RNA was isolated from cells harvested at the indicated time (min) after stop of transcription by rifampin addition to cultures at an OD600 of 0.5. Stabilities of the overproduced rreB derivatives 16c-rreB(GUG/UAG) and 17c-rreB were calculated from 4 biological experiments and are given below the panels.
Similar small genes in other αproteobacteria
Assuming that the 17 nt complementarity to the 3′-end of 16S rRNA is a hallmark of rreB and its functional homologs, we used a 20 nt query corresponding to the 3′-end of 16S rRNA for a BLASTN search in bacterial genomes. We found putative rreB analogs containing a 16 nt or 17 nt region of complementarity to the 3′-end of 16S rRNA in many representatives of Caulobacterales, Rhizobiales, Rhodobacterales, and Rhodospirillales. It is noteworthy that in contrast to other α-proteobacteria, in Azospirillum the rre gene is located on a plasmid and the sORF starts with an ATG. Although the genomic context of rreB (Fig. 1A) is quite conserved in Bradyrhizobiaceae, it is not conserved in other α-proteobacteria.
Fig. 8 shows the conservation of the small genes named rreR in the Rhodobacter/Roseobacter clade. In all cases the last nucleotide of the large complementarity region belongs to a GTG start codon of a sORF comprising 18 codons. There is no sequence similarity between the small proteins RreB and RreR, but they are well conserved in the respective phylogenetic clades (see Fig. 2B, Fig. S1, and Fig. 8B). RreR from Dinoroseobacter shibae DFL12 (18 aa) (Fig. 8C) was also synthesized and analyzed by CD- and 1D-NMR spectroscopy. However, similarly to RreB, RreR remained largely unstructured in solution (Figs S9 to S12).
Figure 8.

Putative rreB homologs in Rhodobacterales. (A) Multiple sequence alignment of sequences corresponding to the putative small mRNAs containing homologous sORFs of 18 codons (named rreR) and preceded by a region with extended complementarity to the 3′-end of 16 sRNA (Extended SD region). Canonical SD, GTG start codon and stop codons are highlighted in gray. For other descriptions see (Fig. 2A. (B) Multiple sequence alignment of the small proteins RreR encoded by the sORFs shown in A). O.ar.238, Octadecabacter arcticus 238; O.an.307, Octadecabacter antarcticus 307; J.sp.1, Jannaschia sp CCS 1; Ros.e.19469, Roseibacterium elongatum DSM 19469; Ro.d.114, Roseobacter denitrificans OCh 114; Ro.l.149, Roseobacter litoralis Och 149; D.s.12, Dinoroseobacter shibae DFL 12; P.t.23, Planktomarina temperata RCA 23; Ph.g.26640, Phaeobacter gallaeciensis DSM 26640; Ph.i.17395, Phaeobacter inhibens DSM 17395; Pa.a.7686, Paracoccus aminophilus JCM 7686; R.c.1003, Rhodobacter capsulatus SB 1003; Pa.d 1222, Paracoccus denitrificans PD 1222; Rh.s.2351, Rhodovulum sulfidophilum DSM 2351; C.i.P73, Celeribacter indicus strain P73; R.s.2.4.1., Rhodobacter spaheroides ATCC 2.4.1. (C) Predicted spatial structure of RreR from D. shibae DFL 12.
Discussion
In this work we identified a small mRNA named rreB with an extraordinary extended SD (17c-region) in B. japonicum and found that it is highly conserved in the family Bradyrhizobiaceae. The conservation of an exceptional SD followed by sORF in many bacteria beyond Bradyrhizobiaceae shows evolutionary pressure for maintenance of such arrangement. However, so far the function of rre genes remains elusive. The similar growth of the EVC and B. japonicum which overproduces the ribosome-binding 17c-rreB mRNA suggests that simple recruitment of ribosomes for preventing translation is not a major function of rreB. This result can be explained by the much higher number of ribosomes when compared with native and overexpressed rreB mRNA, as judged from intensity signals in Northern blot hybridizations (data not shown). Further, putative overproduction of the small protein RreB in the strain harboring pJH-14c did not influence growth, thus preventing further analysis of its physiological role.
The secondary structure prediction of rreB suggested that only the 5 5′ nucleotides of the 17c-region are single-stranded and thus accessible for ribosome binding, while the downstream nucleotides are involved in a conserved hairpin structure (Fig. 2D). We suggest that the predicted single-stranded part of the 17c-region probably functions as a seed region for interaction with 16S rRNA, leading to subsequent melting of the first stem-loop of rreB and binding to the 30S ribosomal subunit. This assumption would explain the efficient binding of rreB and its 17c-rreB derivative to ribosomes (as shown by their predominant localization in the P100 fraction) as well as the much lower proportion of 14c-rreB RNA in P100 (see Fig. 4). The last transcript should harbor only 2 nt in the single-stranded region for seed interaction with 16S rRNA and thus it can be expected that its binding to ribosomes is inefficient.
It is noteworthy that when expressed in E. coli (Fig. 4), rreB was weakly associated with ribosomes (Fig. S6), pointing to differences in the accessibility of its RBS in the 2 bacterial species. Due to small differences between the 16S rRNA sequences of B. japonicum and E. coli, the region of complementarity between E. coli 16S rRNA and rreB is 14 nt with a potential seed interaction region of 3 nucleotides. The latter could be a reason for the inefficient interaction between rreB and ribosomes in E. coli.
A 17 bp duplex formation between rreB and 16S rRNA would encompass the entire single-stranded 3′-end of 16S rRNA in B. japonicum. A previous crystallography study with an artificial RNA has shown that 5 nt downstream of the canonical SD can be involved in the formation of the SD helix, which in that case had a length of 12 bp and involved almost all nucleotides of the single-stranded 3′-end of 16S rRNA of Thermus thermophilus.38 Our data revealed important roles for the 9 5′ nucleotides of the 17c-region: the 5′-extension upstream of the canonical 6 nt SD AGGAGG was crucial for the highly efficient rreB binding to ribosomes (Fig. 4) and the full canonical SD was needed for rreB translation (compare lane 4 to lane 5 in Fig. 5). Furthermore, the data suggest that the 8 3′ nucleotides of the 17c-region (the 7 nt between the canonical SD and the start codon, and the first nucleotide of the start codon) serve to minimize rreB translation: contrary to our expectations, the 11c-egfp mRNA harboring AGG was not translated and the 14c-egfp-mRNA with canonical SD was translated very weakly when compare with the construct with standard SD (Fig. 5).6,10 Finally, the GTG start codon was identified as an additional determinant keeping rreB translation low (Fig. 6C). Thus, our results show that the unique 17c-region enables very efficient binding of rreB to ribosomes but keeps its translation weak. It is not clear whether translation is weak under all conditions or rreB it is efficiently translated under specific, yet unknown conditions.
The rreB stability was dependent on the start codon (Fig. 7), suggesting that rreB degradation depends either on the 30S initiation complex or on the 70S ribosome. We favor the latter possibility, since it is known that mRNA is cleaved in ribosomes stalled due secondary structure roadblocks.39 Strong binding of rreB mRNA to 16S rRNA that prevents elongation would result in a similarly trapped ribosome and may lead to rreB cleavage and ribosome rescue.
We used the unusually extended complementarity of rreB to 16S rRNA to search for similar or similarly regulated genes in other bacteria and detected putative rre genes in many α-proteobacteria. The high similarity between the RBS of rreB and of other α-proteobacterial rre mRNAs suggests strong interaction with ribosomes and similar posttranscriptional regulation at the level of degradation and translation. This shows how features of non-translated regions may help to uncover new genes for small proteins without detectable homology at the amino acid sequence level. The conservation of the rreB sORF in Bradyrhizobiaceae, the rreR sORF in Rhodobacterales and rre sORFs in other α-proteobacterial lineages suggests that these small genes are important for survival of many α-proteobacteria.
Supplementary Material
Author contributions
J.H and S.T. characterized experimentally rreB, E.E.H., A.M. and S.T. performed bioinformatic analyses, R.F.v.B. performed flow cytometry and FACS, N.K. and H.S. performed CD and NMR spectrometry, J.H., E.K., H.S., and E.E.H. wrote the manuscript, E.E.H. conceived the study. All authors read and approved the final manuscript.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgments
We are grateful to Florian Rossmann and Kai Thormann (University of Giessen) for help with fluorescence microscopy, and to Julia Frunzke (Forschungszentrum Jülich) for support with flow cytometry and FACS. We also thank Saina Azarderakhsh and Franziska Kranz (University of Giessen) for help in some experiments and Hans-Martin Fischer (ETH Zürich) for providing soybean nodules.
Funding
This work was supported by the Deutsche Forschungsgemeinschaft (DFG) under grant Ev42/4-2 (to E.E.H.); and the Russian Scientific Foundation under grant N 14-24-00061 (to A. M. and E. K; Fig. 1, Fig. 8 and Fig. S1). J.H. was member of IRTG 1384 “Enzymes and multienzyme complexes acting on nucleic acids” supported by the DFG. Work at BMRZ is supported by state of Hesse. H.S. is member of the DFG-funded cluster of excellence: macromolecular complexes.
References
- 1.Sharma CM, Vogel J. Differential RNA-seq: the approach behind and the biological insight gained. Curr Opin Microbiol 2014; 19:97-105 PMID:25024085; https://doi.org/25890157 10.1016/j.mib.2014.06.010 [DOI] [PubMed] [Google Scholar]
- 2.Prasse D, Thomsen J, De Santis R, Muntel J, Becher D, Schmitz RA. First description of small proteins encoded by spRNAs in Methanosarcina mazei strain Gö1. Biochimie 2015; 117:138-48; PMID:25890157; https://doi.org/ 10.1016/j.biochi.2015.04.007 [DOI] [PubMed] [Google Scholar]
- 3.Storz G, Wolf YI, Ramamurth KS. Small proteins can no longer be ignored. Annu Rev Biochem 2014; 83:753-77; PMID:24606146; https://doi.org/ 10.1146/annurev-biochem-070611-102400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gimpel M, Heidrich N, Mäder U, Krügel H, Brantl S. A dual-function sRNA from B. subtilis: SR1 acts as a peptide encoding mRNA on the gapA operon. Mol Microbiol 2010; 76:990-1009; PMID:20444087; https://doi.org/ 10.1111/j.1365-2958.2010.07158.x [DOI] [PubMed] [Google Scholar]
- 5.Shine J, Dalgarno L. The 3′-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A 1974; 71:1342-6; PMID:4598299; https://doi.org/ 10.1073/pnas.71.4.1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Omotajo D, Tate T, Cho H, Choudhary M. Distribution and diversity of ribosome binding sites in prokaryotic genomes. BMC Genomics 2015; 16:604; PMID:26268350; https://doi.org/ 10.1186/s12864-015-1808-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warren AS, Archuleta J, Feng WC, Setubal JC. Missing genes in the annotation of prokaryotic genomes. BMC Bioinformatics 2010; 11:131; PMID:20230630; https://doi.org/ 10.1186/1471-2105-11-131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuroda H, Suzuki H, Kusumegi T, Hirose T, Yukawa Y, Sugiura M. Translation of psbC mRNAs starts from the downstream GUG, not the upstream AUG, and requires the extended Shine-Dalgarno sequence in tobacco chloroplasts. Plant Cell Physiol 2007; 48:1374-78; PMID:17664183; https://doi.org/ 10.1093/pcp/pcm097 [DOI] [PubMed] [Google Scholar]
- 9.Komarova AV, Tchufistova LS, Supina EV, Boni IV. Protein S1 counteracts the inhibitory effect of the extended Shine-Dalgarno sequence on translation. RNA 2002; 8:1137-47; PMID:12358433; https://doi.org/ 10.1017/S1355838202029990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osterman IA, Evfratov SA, Sergiev PV, Dontsova OA. Comparison of mRNA features affecting translation initiation and reinitiation. Nucleic Acids Res 2013; 41:474-86; PMID:23093605; https://doi.org/ 10.1093/nar/gks989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ramamurthi KS, Storz G. The small protein floodgates are opening; now the functional analysis begins. BMC Biol 2014; 12:96; PMID:25475548; https://doi.org/ 10.1186/s12915-014-0096-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tenson T, DeBlasio A, Mankin A. A functional peptide encoded in the Escherichia coli 23S rRNA. Proc Natl Acad Sci U S A 1996; 93:5641-6; PMID:8643630; https://doi.org/ 10.1073/pnas.93.11.5641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edwards A, Frederix M, F Wisniewski-Dyé, Jones J, Zorreguieta A, Downie JA. The cin and rai quorum-sensing regulatory systems in Rhizobium leguminosarum are coordinated by ExpR and CinS, a small regulatory protein coexpressed with CinI. J Bacteriol 2009; 191:3059-67; PMID:19270098; https://doi.org/ 10.1128/JB.01650-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vogel J, Argaman L, Wagner EG, Altuvia S. The small RNA IstR inhibits synthesis of an SOS-induced toxic peptide. Curr Biol 2004; 14:2271-6; PMID:15620655; https://doi.org/ 10.1016/j.cub.2004.12.003 [DOI] [PubMed] [Google Scholar]
- 15.Fozo EM, Kawano M, Fontaine F, Kaya Y, Mendieta KS, Jones KL, Ocampo A, Rudd KE, Storz G. Repression of small toxic protein synthesis by the Sib and OhsC small RNAs. Mol Microbiol 2008; 70:1076-93; PMID:18710431; https://doi.org/ 10.1111/j.1365-2958.2008.06394.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lewis K. Persister cells. Annu Rev Microbiol 2010; 64:357-72; PMID:20528688; https://doi.org/ 10.1146/annurev.micro.112408.134306 [DOI] [PubMed] [Google Scholar]
- 17.Desbrosses GJ, Stougaard J. Root nodulation: a paradigm for how plant-microbe symbiosis influences host developmental pathways. Cell Host Microbe 2011; 10:348-58; PMID:22018235; https://doi.org/ 10.1016/j.chom.2011.09.005 [DOI] [PubMed] [Google Scholar]
- 18.Fischer HM. Genetic regulation of nitrogen fixation in rhizobia. Microbiol Rev 1994; 58:352-86; PMID:7968919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Čuklina J, Hahn J, Imakaev M, Omasits U, Förstner KU, Ljubimo N, Goebel M, Pessi G, Fischer HM, Ahrens CH, et al.. Genome-wide transcription start site mapping of Bradyrhizobium japonicum grown free-living or in symbiosis - a rich resource to identify new transcripts, proteins and to study gene regulation. BMC Genomics 2016; 17:302; PMID:27107716; https://doi.org/ 10.1186/s12864-016-2602-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaneko T, Nakamura Y, Sato S, Minamisawa K, Uchiumi T, Sasamoto S, Watanabe A, Idesawa K, Iriguchi M, Kawashima K, et al.. Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res 2002; 9:189-97; PMID:12597275; https://doi.org/ 10.1093/dnares/9.6.189 [DOI] [PubMed] [Google Scholar]
- 21.Regensburger B, Hennecke H. RNA polymerase from Rhizobium japonicum. Arch Microbiol 1983; 135:103-9; PMID:6639271; https://doi.org/ 10.1007/BF00408017 [DOI] [PubMed] [Google Scholar]
- 22.Delamuta JR, Ribeiro RA, Ormeño-Orrillo E, Melo IS, Martínez-Romero E, Hungria M. Polyphasic evidence supporting the reclassification of Bradyrhizobium japonicum group Ia strains as Bradyrhizobium diazoefficiens sp. nov. Int J Syst Evol Microbiol. 2013; 63:3342-51; PMID:23504968; https://doi.org/ 10.1099/ijs.0.049130-0 [DOI] [PubMed] [Google Scholar]
- 23.Mesa S, Hauser F, Friberg M, Malaguti E, Fischer HM, Hennecke H. Comprehensive assessment of the regulons controlled by the FixLJ-FixK2-FixK1 cascade in Bradyrhizobium japonicum. J Bacteriol 2008; 190:6568-79; PMID:18689489; https://doi.org/ 10.1128/JB.00748-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 1985; 33:103-19; PMID:2985470; https://doi.org/ 10.1016/0378-1119(85)90120-9 [DOI] [PubMed] [Google Scholar]
- 25.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Biotechnology 1982; 1:784-91; https://doi.org/ 10.1038/nbt1183-784 [DOI] [Google Scholar]
- 26.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 1989. [Google Scholar]
- 27.Alvarez-Morales A, Betancourt-Alvarez M, Kaluza K, Hennecke H. Activation of the Bradyrhizobium japonicum nifH and nifDK operons is dependent on promoter-upstream DNA sequences. Nucleic Acids Res 1986; 14:4207-27; PMID:3086837; https://doi.org/ 10.1093/nar/14.10.4207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rudolph G, Semini G, Hauser F, Lindemann A, Friberg M, Hennecke H, Fischer HM. The Iron control element, acting in positive and negative control of iron-regulated Bradyrhizobium japonicum genes, is a target for the Irr protein. J Bacteriol 2006; 188:733-44; PMID:16385063; https://doi.org/26035130 10.1128/JB.188.2.733-744.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ledermann R, Bartsch I, Remus-Emsermann MN, Vorholt JA, Fischer HM. Stable Fluorescent and Enzymatic Tagging of Bradyrhizobium diazoefficiens to Analyze Host-Plant Infection and Colonization. Mol Plant Microbe Interact 2015; 28:959-67; PMID:26035130; https://doi.org/ 10.1094/MPMI-03-15-0054-TA [DOI] [PubMed] [Google Scholar]
- 30.Madhugiri R, Pessi G, Voss B, Hahn J, Sharma CM, Reinhardt R, Vogel J, Hess WR, Fischer HM, Evguenieva-Hackenberg E. Small RNAs of the Bradyrhizobium/Rhodopseudomonas lineage and their analysis. RNA Biol 2012; 9:47-58; PMID:22258152; https://doi.org/19800865 10.4161/rna.9.1.18008 [DOI] [PubMed] [Google Scholar]
- 31.Madhugiri R, Evguenieva-Hackenberg E. RNase J is involved in the 5′-end maturation of 16S rRNA and 23S rRNA in Sinorhizobium meliloti. FEBS Lett 2009; 583:2339-42; PMID:19540834; https://doi.org/19800865 10.1016/j.febslet.2009.06.026 [DOI] [PubMed] [Google Scholar]
- 32.Voss B, Hölscher M, Baumgarth B, Kalbfleisch A, Kaya C, Hess WR, Becker A, Evguenieva-Hackenberg E. Expression of small RNAs in Rhizobiales and protection of a small RNA and its degradation products by Hfq in Sinorhizobium meliloti. Biochem Biophys Res Commun 2009; 390:331-6; PMID:19800865; https://doi.org/ 10.1016/j.bbrc.2009.09.125 [DOI] [PubMed] [Google Scholar]
- 33.Rheinberger HJ, Geigenmüller U, Wedd M, Nierhaus KH. Parameters for the preparation of Escherichia coli ribosomes and ribosomal subunits active in tRNA binding. Methods Enzymol 1988; 164:658-70; PMID:3071687; https://doi.org/ 10.1016/S0076-6879(88)64076-6 [DOI] [PubMed] [Google Scholar]
- 34.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 1990; 215:403-10; PMID:2231712; https://doi.org/ 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- 35.Smith C, Heyne S, Richter AS, Will S, Backofen R. Freiburg RNA Tools: a web server integrating INTARNA, EXPARNA and LOCARNA. Nucleic Acids Res 2010; 38:W373-7; PMID:20444875; https://doi.org/ 10.1093/nar/gkq316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al.. Clustal W and Clustal X version 2.0. Bioinformatics 2007; 23:2947-48; PMID:17846036; https://doi.org/ 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 37.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 2015; 10:845-58; PMID:25950237; https://doi.org/ 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yusupova G, Jenner L, Rees B, Moras D, Yusupov M. Structural basis for messenger RNA movement on the ribosome. Nature 2006; 444:391-4; PMID:17051149; https://doi.org/ 10.1038/nature05281 [DOI] [PubMed] [Google Scholar]
- 39.Liang W, Rudd KE, Deutscher MP. A role for REP sequences in regulating translation. Mol Cell 2015; 58:431-9; PMID:25891074; https://doi.org/10518617 10.1016/j.molcel.2015.03.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zahn K, Inui M, Yukawa H. Characterization of a separate small domain derived from the 5′ end of 23S rRNA of an alpha-proteobacterium. Nucleic Acids Res 1999; 27:4241-50; PMID:10518617; https://doi.org/ 10.1093/nar/27.21.4241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Evguenieva-Hackenberg E. Bacterial ribosomal RNA in pieces. Mol Microbiol 2005; 57:318-25; PMID:15978067; https://doi.org/ 10.1111/j.1365-2958.2005.04662.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


