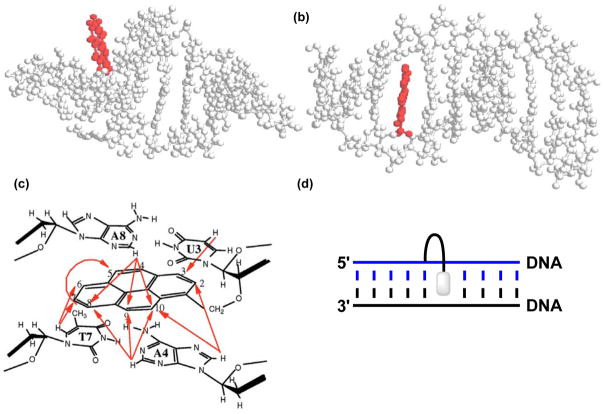
Figure 2.
Structures obtained from MD simulations of a U-modified (a) RNA duplex (5′-r-CCUCAUGAGG-3′ and cRNA) and (b) DNA duplex (5′-d-CCUCATGAGG-3′ and cDNA). (c) Observed NOE contacts between nucleobase and pyrene protons in a U-modified DNA duplex (5′-C1C2U3A4G5C6T7A8G9G10 : 3′-G10G9A8T7C6G5A4U3C2C1). (d) Illustration of a 3′-intercalative binding mode. Panels a–c are reproduced from reference 19 with permission from Oxford University Press.