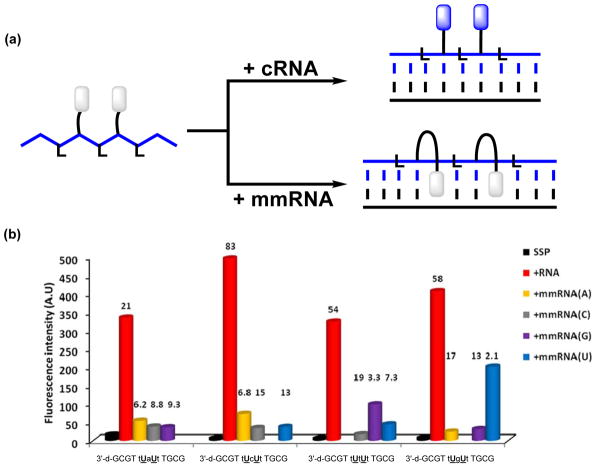
Figure 4.
(a) DNA probes with central segments of alternating LNA and U-monomers result in large emission enhancements upon binding with cRNA, whereas duplexes with mismatched RNA targets only are scarcely emissive. (b) Fluorescence intensity at λem = 376 nm of single-stranded probes (SSP) and duplexes with complementary or centrally mismatched RNA targets (mmRNA, nature of mismatched nucleotide is specified) (λex = 350 nm). Lower case a, c, g and t denote LNA monomers. Hybridization-induced increases (i.e., the intensity ratio between a matched duplex and single-stranded probe) and discrimination factors (i.e., the intensity ratio between a matched and mismatched duplex) are listed above the corresponding histograms. Adapted from reference 27 with permission from the Royal Society of Chemistry.