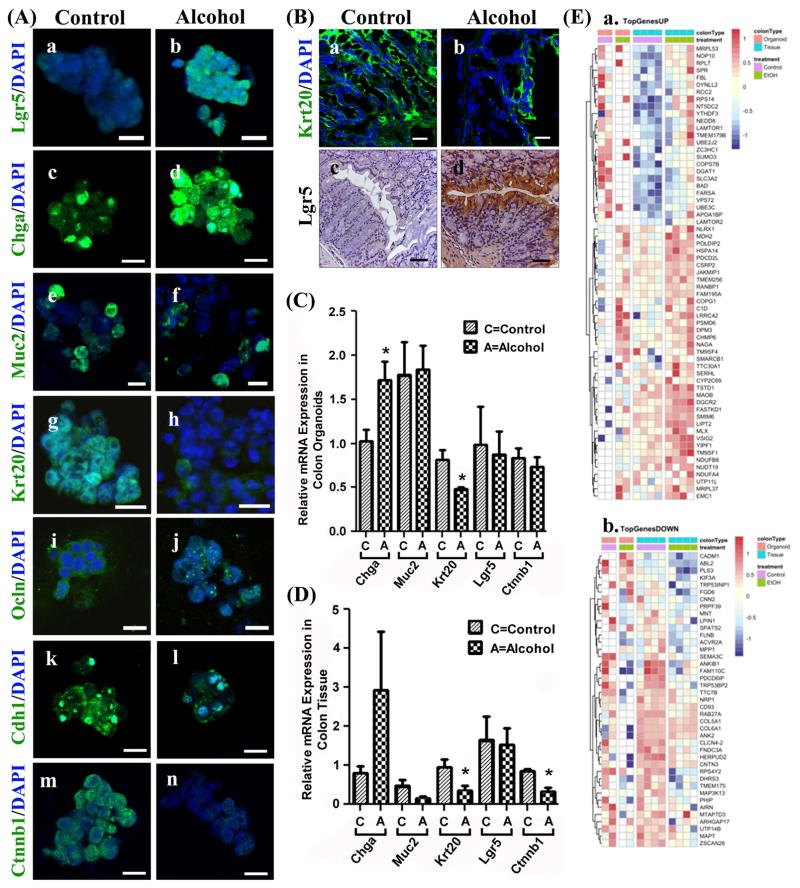
Figure 4. Colonic organoids and tissue from alcohol fed mice exhibit differences in cell lineage markers, apical junction proteins, stem cell markers and RNA-seq.
(A.) Organoids grown from colonic intestinal tissue from control (Control) or alcohol-fed (Alcohol) mice were stained for cell lineage and other key markers (Methods): (a/b) Lgr5; (c/d) Enteroendocrine cells: chromogranin A (Chga); (e/f) Goblet cells: mucin 2 (Muc2); (g/h) Colonocytes/enterocytes: cytokeratin 20 (Krt20); (i/j) tight junction protein occludin (Ocln); (k/l) E-cadherin (Cdh1); and (m/n) β-catenin (Ctnnb1). (B.) Whole intestinal tissue from colons of Control (left) or Alcohol fed (right) mice were stained for (a/b) cytokeratin 20 (Krt20)(green) or (c/d) Lgr5 (DAB staining, brown) as described in Methods. Tissue from colon organoids (C.) or colon tissue (D.) were analyzed by qPCR (Methods) for gene expression including Chga, Muc2, Krt20, Lgr5, and Ctnnb1(*p<.05)(t-test) (E.) RNA-seq analysis data from colon tissue and organoids from control and alcohol fed mice was used to construct heat maps to depict differential gene expression with alcohol feeding (Methods). (a.) TopGenesUP (43 genes) and (b.)TopGenesDOWN (33 genes): Heat maps of genes from colon samples that are common between organoids and colon tissue that are upregulated (TopgenesUP) or downregulated (TopGenesDOWN) in alcohol fed mice. LogCPM data was z-scored transformed and plotted using the Bioconductor pheatmap.(Kolde, 2015) Genes were clustered by complete linkage method using Euclidean distance. The analysis was performed for the contrast of alcohol fed vs. control, and so the red color indicates increased expression in alcohol fed mice, while blue indicates increased expression in control mice. The first colored row at the top of the plot indicate the type of colon sample: orange is organoid cells, blue is tissue. The second colored band indicates the treatment: green are alcohol fed samples while purple are controls. Scale bars in A (a–n) represent 10μm and in D (a, b) represent 20μm and D (c, d) represent 50μm.