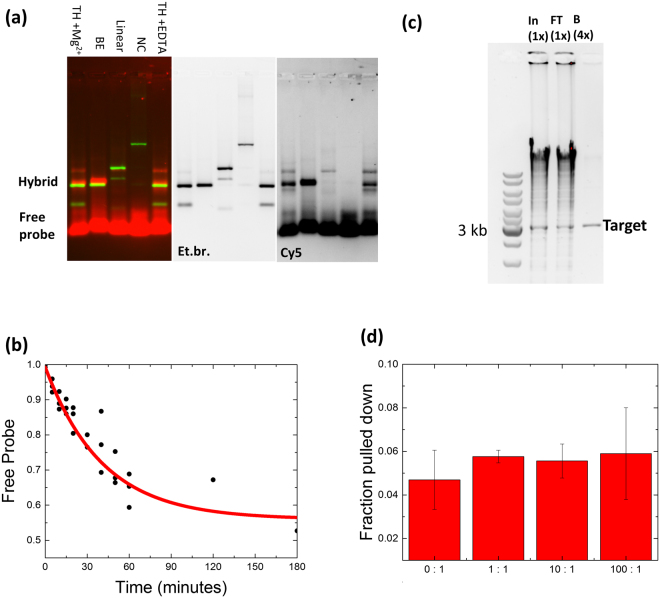
Figure 2.
Sequence specific hybridization of an LNA probe with the target sequence. (a) The LNA probe has increased binding for a target with a compatible toehold. The Cy5 labeled pLNA-1 was hybridized to different targets, TH: target with toehold, in the presence of either 10 mM Mg2+ or 10 mM EDTA, BE: blunt end target including toehold sequence, Linear: DNA fragment with the target sequence at 116 bp from the end; NC: DNA fragment without the target sequence (Sequences in Supplementary Table S1). After hybridization the DNA was separated in a 2% agarose gel and visualized with Ethidium bromide (EtBr, green) and the Cy5 label (red) (left panel, composite). The separate Ethidium bromide and Cy5 channels are shown on the right. (b) Hybridization of the LNA probe (5.6 nM) and target (56 nM) over time, measured by gel electrophoresis (see Figure S1). Data was fitted with a single exponential decay. The offset corresponds to Kd of 39 nM, yielding a half-life for the unbound probe of 60 minutes. (c) Genomic DNA from E.coli digested with HindIII was mixed with target DNA in a 100:1 ratio, and subjected to purification using an LNA toehold as described in Methods. DNA contained in samples from input (In), flow through (FT), and recovered from beads (B) was separated in an agarose gel and visualized with EtBr. The relative fraction of each sample loaded is indicated on top. Positions of selected marker bands in the first lane and the target are shown on the left and right, respectively. (d) Quantification of the recovery of the DNA target from different mixtures with varying competitor (genomic DNA from E. coli) to target ratios.