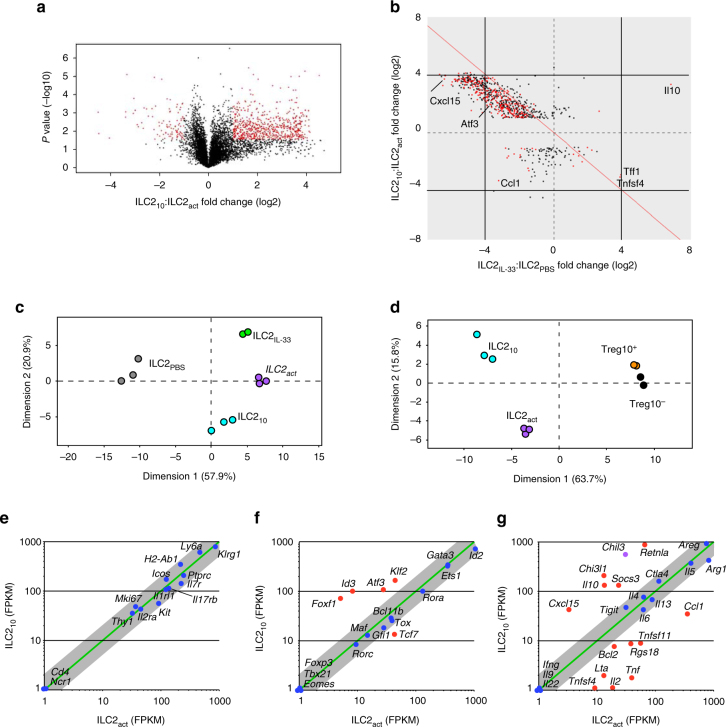
Fig. 3.
ILC210 represent a molecularly distinct ILC2 activation state. a Volcano plot for differentially expressed genes, defined as in Fig. 1, comparing ILC210 and IL-33 activated IL-10– ILC2 (ILC2act). Genes defined as statistically significant analyzed as in Fig. 1 are shown in red. b Fold-change plot of differentially expressed genes in the intersection of indicated ILC2 populations. Genes defined as statistically significant in a are shown in red. c Principal component analysis of RNA-seq data from purified total ILC2 from PBS (ILC2PBS) or IL-33 (ILC2IL-33) injected mice, or from isolated ILC210 or ILC2act from IL-33 injected mice. d Principal component analysis of RNA-seq data comparing ILC210 and ILC2act, with IL-10+ and IL-10– Treg cells. e–g Selected genes plotted as the average FPKM from ILC210 vs. ILC2act, presented as in Fig. 1. Shown are expression of selected genes grouped by (e) ILC2 markers and markers of activation, (f) transcription factors, and (g) effector molecules (Chil3, purple, P = 0.05). Data are from two or three independent experiments with five mice pooled per experiment a–g