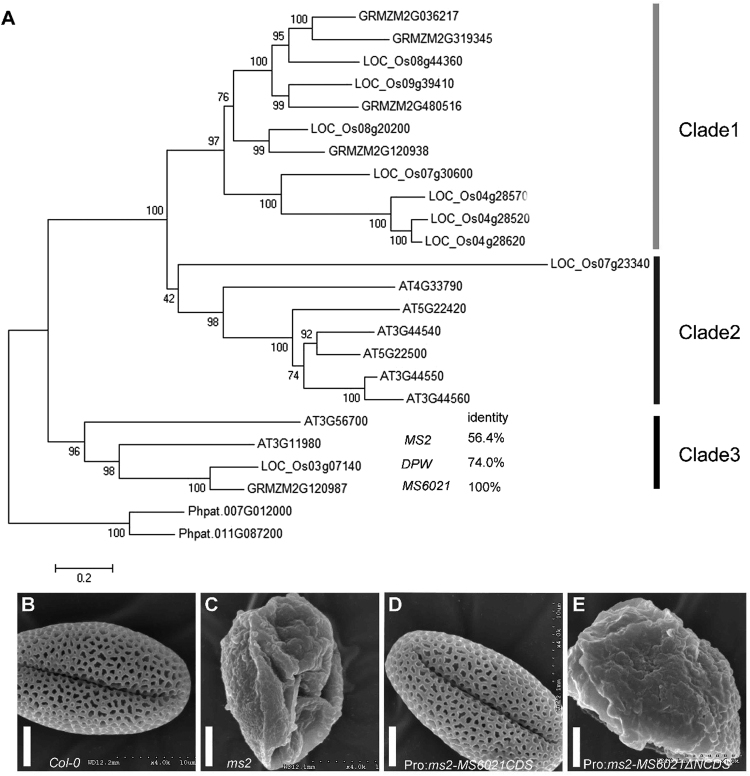
Figure 8.
Functional conservation of MS6021. (A) A neighbor-joining phylogenetic tree summarizing the evolutionary relationships among FAR members in Arabidopsis, rice, and maize of (BLASTPE E-value < 1E-100). The proteins are named according to their Phytozome accession numbers. The numbers under the branches refer to the bootstrap value of the neighbor-joining phylogenetic tree. The length of the branches is proportional to the amino acid variation rates. At, Arabidopsis thaliana; Os, Oryza sativa; Zm, Zea mays. The bar indicates the estimated number of amino acid substitutions per site (for the protein alignment, see Figure S4). (B) to (E) SEM analysis of the pollen wall surface of wild-type (B), ms2 mutant (C), and transgenic ms2 mutant lines (D and E) at anthesis. Bars = 5 µm.